Volume 10, Issue 1 (2022)
Health Educ Health Promot 2022, 10(1): 193-200 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Obeed F, Al-Saad L, Alrubayae I. The Prevalence of Hospital-Acquired Fungal Infections in Some Hospitals of Basrah Province. Health Educ Health Promot 2022; 10 (1) :193-200
URL: http://hehp.modares.ac.ir/article-5-59900-en.html
URL: http://hehp.modares.ac.ir/article-5-59900-en.html
1- Department of Biology, College of Science, University of Basrah, Basrah, Iraq
2- College of Pharmacy, University of Basrah, Basrah, Iraq
2- College of Pharmacy, University of Basrah, Basrah, Iraq
Full-Text [PDF 929 kb]
(3596 Downloads)
| Abstract (HTML) (1683 Views)
Full-Text: (607 Views)
Introduction
A hospital-acquired infection (HAI), also known as a nosocomial infection, is a form of infection that occurs in hospitals. The World Health Organization (WHO) describes HAIs as infections obtained while the patient is hospitalized but not present or incubating at the time of admission. The infections that occur more than 48 hours after admission to the hospitals are of big concern in the healthcare system, they are a leading cause of preventable disease. It has detrimental implications on both medical conditions and institutional efficiencies, including longer hospital visits, long-term injury, increased microorganism tolerance, substantial financial pressure (both patient and institution), and excess mortality [1, 2].
Nosocomial fungal infections are amongst the main causes of mortality in patients admitted to healthcare settings, especially in immunocompromised populations. The predominant pathogens include Candida spp., Aspergillus spp., Mucorales spp., and Fusarium spp. [3].
Over the last decades, the prevalence of invasive fungal infection has gradually increased, though, with a recent medical and surgical advancements, interventions, and an ever-growing population, the diversity and immunocompromised patients, also the number of fungal pathogens continues to increase, like Candida spp. are opportunistically invasive in people whose immune systems are weak [4, 5].
The risk and severity of infection increase in some special units in hospitals, such as intensive care units, child care units, burns, and units for patients with weak immunity [2].
Candida spp., especially C. albicans, are part of the human microbial flora; hence, most Candida infections are endogenous in origin. Candidemia disseminated hematogenous infections, or deep-seated infections can occur in immunocompromised patients, such as those with neutropenia, and critically ill patients [6].
In patients with chemotherapy-induced neutropenia and mucositis, candidemia may originate from the gastrointestinal tract. However, in critically ill patients, the source of candidemia is most likely a central vascular catheter (CVC) colonized by Candida spp. from the patient’s endogenous microflora or acquired from the health care environment [7].
Candida spp. have been isolated from environmental cultures of the floor, countertops, and other inanimate surfaces in the hospital. Patient acquisition and colonization with Candida spp. found in the hospital environment and food has been demonstrated. The propensity of Candida spp., especially C. parapsilosis, to cause Central line-associated bloodstream infections (CLABSIs) is likely related to this pathogen’s ability to form biofilms on catheters [3, 7].
Nosocomial infections are most difficult to diagnose, so early empiric therapy for patients who are clinically suspected of having a fungal infection, or prophylaxis for the highest-risk individuals, has been prioritized [4].
According to the scarcity of studies about nosocomial fungal infection and hospital community fungal profiling in Iraq, the current study was aimed at the prevalence of Hospital-Acquired Fungal Infections in Some Hospitals in Basrah Province.
Material and Methods
Two hundred thirteen clinical specimens were collected from patients after 48h of admission in Al–Faehaa Educational Hospital, AL-Sader Educational Hospital, and Pediatric Specialist Educational Hospital from November 2020 to April 2021, the age of patients ranged from 5 months to 80 years, 94 (44%) males and 119 (56%) females. The clinical specimens were sorted into 59 samples of midstream urine, 56 samples of wound swab, 29 samples of sputum, 27 samples of pleural fluid, 25 samples of blood, 13 samples of fluid, and 4 samples of pus. All clinical specimens were examined microscopically using the 10% KOH and Gram stain technique to show yeast cells and /or pseudohyphae or true hyphae and cultured on SDA with chloramphenicol and blood agar plates, the cultures were incubated at 37 °C for 1-14 days under aerobic conditions.
Yeast isolates were activated on SDA for 1-3 days to evaluate their ability of germ tube formation by adding a small portion of the activated colony to 0.5ml of blood serum. After that, the tubes were incubated at 37°C for 3 hours. The samples were investigated after the incubation period by adding a drop from inoculum on a clean slide and covered with a coverslip, then examined microscopically on 10X and 40X to detect the formation of the elongated tube without constriction from mother cells that indicated germ tube formation [8, 9].
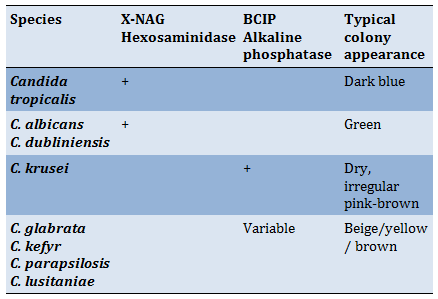
Activated yeast isolates (1-3 days on SDA) were grown on Brilliance TM Candida Agar and incubated at 30°C for 1-3 days to be identified according to their colors and appearance on this test medium according to the following Table 1.
All yeast isolates were diagnosed by Vitek®2 system 7.01 and 8.01 software (BIOMERIEUX, USA) according to the manufacturer's instructions.
A molecular study was performed to confirm the identity of the clinical yeast isolates according to Mirhendi et al. [10]. Genomic DNA was extracted according to the instructions provided by the manufacturer of Presto Mini gDNA yeast kit (Geneaid, Taiwan) after cultivation of a single colony on SDA for 48h at 25ᵒC. The large subunit 28S region of rDNA was considered to identify the isolated fungi using the universal primers Null1 F-5’- GCA TAT CAA TAA GCG GAG GAA AAG -3’ and Null4 R-5’- GGT CCG TGT TTC AAG ACG G -3’. The PCR mixture was prepared in a total volume of 25μl contained 12.5μl Master Mix (Bioneer, Korea), 1.5μl of each primer,1.0μl of genomic DNA as a template, and 9μl of nuclease-free water. The reaction was performed using (Applied Bio System, USA) thermal cycler and the PCR conditions were 95oC for 1 min followed by 35 cycles of 95ᵒC for 1 min, 56ᵒC for 45 sec, and 72ᵒC for 1 min followed by 1 min of final extension at 72ᵒC. The PCR product was visualized on 2% agarose gel electrophoresis, the gel was pre-stained with ethidium bromide. The PCR product (20μl) of each sample was sent to Macrogen Company (Korea) for purification and sequencing, the sequences were processed by Chromas 2.6.6 and then identified using Basic Local Alignment Search Tool (BLAST) for multiple comparisons with deposited reference strains of National Center of Biotechnology Information (NCBI).
All statistical analysis was performed using Statistical Package for the Social Sciences (SPSS) version 24. The non-parametric variable was compared using the Chi-square for bivariate, while Kruskal-Wallis Test was implemented to analyze multiple comparisons of non-parametric variables.
Table 1) Expected reaction on Brilliance TM Candida Agar
Findings
The results showed that nosocomial fungi were isolated from 27.7% of the total collected specimens. The heights prevalence of fungal infections according to age group was recorded in 21-30 age group which is significantly exceeded the other groups (p<0.05), as well as the result was explained that female shown high prevalence (57.62%) of fungal infections, which significantly differed than males (p<0.05; Table 2).
Figure 1 illustrated that the diabetic patients tended to be significantly (p<0.05) more susceptible to fungal infections than other patients (43.7%), while 30% of the malignant patients were infected.
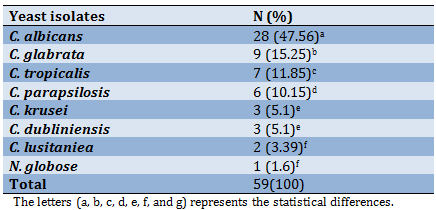
A total of 59 yeast species belonging to two genera were identified from HAFIs after the incubation period. C. albicans was recorded as the most common species 47.56%, followed by C. glabrata at 15.25%, while Naganishia globose appeared in one blood specimen at 1.6% (Table 3).
Table 2) Prevalence of nosocomial fungal infections according to age groups and sex
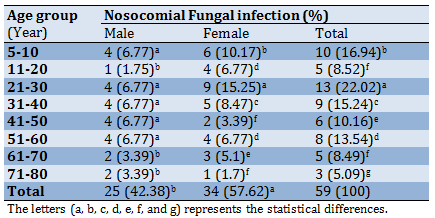
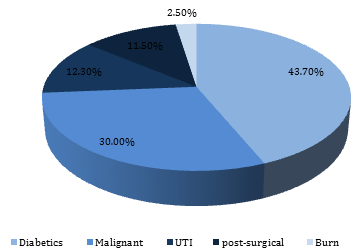
Figure 1) The fungal infections prevalence according to the presence of chronic diseases
The microscopic examination (Figure 2) determined that nosocomial fungal infection was concentrated in the midstream urine specimens (50%) which surpassed both pus and wound significantly (p=0.034) with no significant differences among all other specimens comparison.

Figure 2) The percentage of positive specimens by microscopic direct examination
The microscopic direct examination for clinical specimens Figure 3 (A) represented an aggregation of budding yeast cells in the sputum of a 56 years female patient who spent 3 months in the hospital for coronavirus medication, she had hypoxemia symptoms, so she was transferred to internal medicine ward, Whereas Figure 3 (B) was determined the appearance of true hyphae in wound swab gotten from female post-cesarean surgery patient.
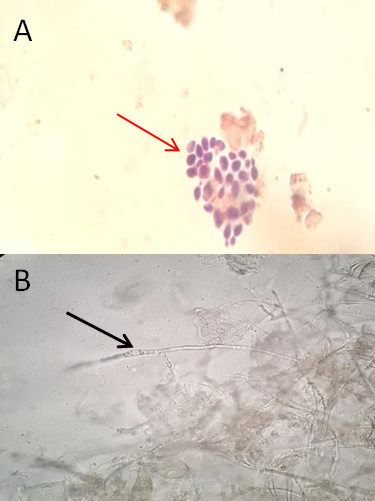
Figure 3) Microscopic direct examination (40X): A- dry film (gram stain) of sputum appears yeasts body; B- wet film (10KOH) of wound swab showing the true hyphae
A total of 59 yeast species belonging to two genera were identified from HAFIs after the incubation period (Table 3). C. albicans (47.56%) was exceeded all other species significantly (p<0.05), followed by Candida glabrata (15.25%) then C. tropicalis with significant differences between them (p=0.05), while the less prevalence recorded in C. lusitaniea (3.39%) and N. globose (1.6%) respectively with no significant differences among them (p=0.157).
Table 3) Prevalence of clinical yeast isolation from HAFIs
Thirty-one (50%) yeast isolates exhibited the ability to generate germ tubes, as shown in Figure 4 the germ tube was determined as free of constriction tube from the mother cell, which is distinguished C. albicans and related species from other Candida spp. as all of these isolates were presumptively identified as C. albicans or C. dubliniensis.
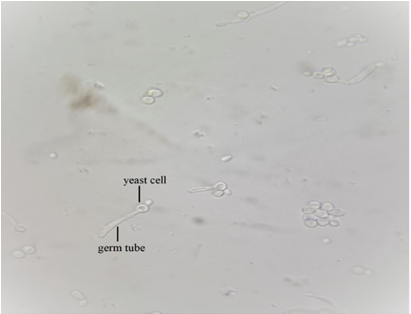
Figure 4) Germ tube formation of C. albicans after 3h incubation at 37oC in human serum (40x)
The differential brilliance Candida agar results (Figure 5) revealed that 59 isolates were returned to the genus Candida: C. albicans, C. dubliniensis, C. glabrata, C. lusitaniea, C. parapsilosis, C. tropicalis, and C. krusei.

Figure 5) Identification of Candida spp. according to the color of colonies on Brilliance Candida agar 1,2,3,4 C. albicans; C. dubliniensis (green color); 5,6 C. glabrata, C. parapsilosis; C. lusitaniea (brown/ beige); 7 C. tropicalis (dark blue).
The results of the Vitek diagnosis (Table 4) showed that C. albicans was the high prevalent species (47.5%), which was significantly surpass the other species (p<0.05) followed by Stephanoascus ciforrii (21.5%), C. tropicalis (15.7%), C. glabrata (7.8%), C. parapsilosis (5.9%), while the lowest prevalence was recorded in C. famata, Trichosporon asahi, C. lusitaniea (3.9%), and C. dubliniensis (2%) with no significant differences (p=0.157).
Table 4) The prevalence of isolated fungi according to the Vitek system diagnosis

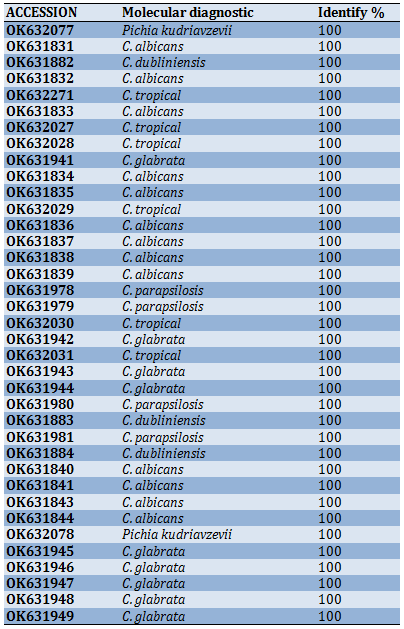
Molecular diagnosis confirmed the identity of the yeast isolates. The sequence analysis of the large subunit of the rDNA (LSU) region that 37 isolates were identified as a Candida spp., which were deposited in the national center for biotechnology information (NCBI) under the accession numbers listed in Table 5.
Table 5) New recording isolates in gen bank
On the other hand, one yeast isolate was identified as N. globose A56_NL which was documented as a new record for the first time in Iraq and deposited in NCBI under the accession number OK597185. Relatively, the new recorded N. globose was isolated from the blood specimens of a 72-years old female patient lying in the burn unit who died after one week of her admission. The heavy growth appeared on blood agar and SDA after 4 days of incubation at 37⁰C as white, dry, irregular colonies, the microscopic examination showed that the yeast cells had broad true hyphae (Figure 6).
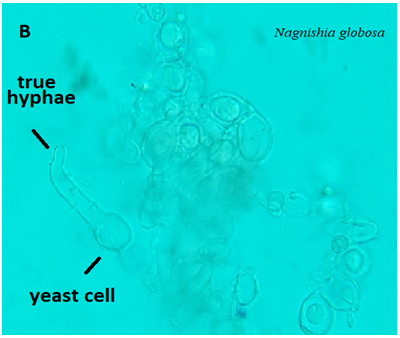
Figure 6) Microscopic examination of N. globose showed that yeast cells and true hyphae (40X)
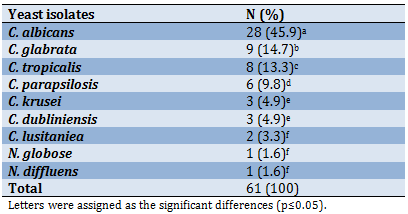
Furthermore, the results of molecular identification (Table 6) revealed that 98.4% of yeast isolates were identified as a Candida spp., among them C. albicans (45.9%) had the superiority over the other species with a significant difference (p<0.05) followed by C. glabrata (14.7%), while C. lusitaniea, N. globose, and N. diffluens recorded the lowest prevalence with no significant differences among them (p=0.157).
Table 6) The prevalence of yeast isolates due to molecular diagnosis
The neighbor-joining tree represented the clustering of the tested isolates according to the related species, which is reflects the sequence homogeneity of the related individuals and the sensitivity of the 28S region of diagnose the fungi to the species level efficiently as it seems that this region is polymorphic enough to distinguish among species and could consider as a good marker for fungi molecular identification. Alternatively, three isolates deviated from this settlement, namely A13, A18, and A30 belonging to three different species that include C. albicans, C. tropicalis, and C. parapsilosis respectively (Figure 7). Also, it illustrated the relationship between one-way sequences (non-assembled, forward, or reverse only) for the same region (28S), and the results revealed the grouping of C. albicans isolates (A4, A11, A12, A14, A15, A17, A20, A27) in one cluster, whilst, the isolates A8 and A15 of C. parapsilosis and C. tropicalis, as well as isolates A7, A16, and A9 of C. albicans and C. parapsilosis were isolated in independent clusters. The cluster pattern here is similar to that of assembled sequences with less accuracy as the one-way sequence mostly being shorter with more noise on both edges, which leads to losing valuable pieces of polymorphic sectors (Figure 8). The results of both trees may recommend considering the assembled sequence for the 28S marker as it seemed that the polymorphic area of that region is relatively narrow.
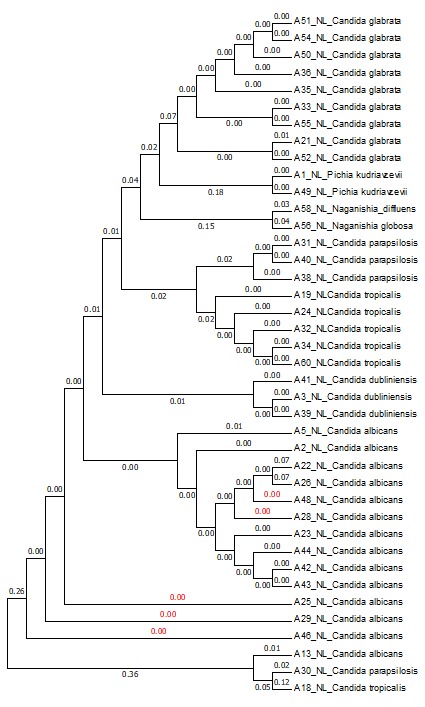
Figure 7) Neighbor-joining tree of the 28S region of assembled sequences. The numbers represent the genetic distance among the isolates

Figure 8) Neighbor-joining tree of Non-Assembled sequences of the 28S region. The numbers represent the genetic distance among the isolates
The results of the molecular method were relatively similar to brilliance candida agar, while the Vitek diagnosis showed inconsistency in 30% of results (Table 7) because the vitek diagnosis depends on the stored database of it that requires updating each six months interval.
Table 7) Comparison among brilliance test, VitekR2 assay, and molecular method

Discussion
The diabetic patients were tend to be significantly (p<0.05) more susceptible to fungal infections than other patients (43.7%), while 30% of the malignant patients were infected, this result was agreement with Al-Duboon [11] and Alrubayae et al. [12]. This is suggests that the high blood sugar can decrease its supplement to the cells which leads to cell weakness of the phagocytes that triggers the virulence of opportunistic microorganisms, including fungi.
The total of 59 yeast species belonging to two genera were identified from HAFIs after incubation
period. C. albicans was recorded as the most common species 47.56%, followed by C. glabrata 15.25%, while N. globose was appeared in one blood specimen with 1.6%. The finding of current study was agreed with other researches that showed C. albicans as the first fungal causative agent caused opportunistic infections in immunocompromised patients [12-15].
The total of 59 yeast species belonging to two genera were identified from HAFIs after incubation period. C. albicans (47.56%) was exceeded all other species, followed by C. glabrata (15.25%) then C. tropicalis, while the less prevalence recorded in C. lusitaniea (3.39%) and N. globose (1.6%) respectively. Similarly, previous studies showed that C. albicans was the most common fungal agent that caused opportunistic infections in immunocompromised patients [12-15]. The ability of C. albicans to affect divers host functions is indeed could be associated with wide range of pathogenic factors and physiological characteristics such as immediate adaptation to pH variation, metabolic adaptability, powerful nutrient acquisition systems and vigorous stress response machineries [16].
Thirty one (50%) yeast isolates exhibited the ability to generate germ tubes. The germ tube was determined as free of constriction tube from the mother cell, which is distinguished C. albicans and related species from other Candida spp. as all of these isolates were presumptively identified as C. albicans or C. dubliniensis. It has become clear that the physiological conditions of immunocompromised patients motivate the dimorphism in C. albicans from budding yeast to hyphal form, therefore, the growth conditions are induced in the laboratory whereby C. albicans grows as yeast only or produce the elongated tube (hyphal state) [17]. These result agreed with Alrubayae et al. [12] and Abbas et al. [15] who reported that almost tested isolates has the ability to form germ tube, which plays an important role of pathogenicity by assistants the pathogen to penetrate host cells barriers for nutrition acquisition [18]. Furthermore, the ability of most isolates to produce other virulence factors that increased their pathogenicity and facilitate penetration and adherence of host tissues like secreted hydrolytic enzymes (proteinase, phospholipase and hemolysis) and biofilm production [19].
The differential brilliance Candida agar results revealed that 59 isolates were returned to the genus Candida. The differential brilliance Candida agar medium mostly considered as primary assay to diagnose and could be differentiate among Candida species [10, 12, 13]. The results of molecular method were relatively similar to brilliance candida agar, while the Vitek diagnosis showed inconsistency in 30% of results, because the vitek diagnoses depends on the stored database of it that requires updating each six months interval, this is agreed with a previous findings performed by Abu-Mejdad et al. [20] and Alrubayae et al. [12].
Conclusion
The current study showed that fungi have an important role as etiological agents of hospital-acquired infections; so, it needs more attention from health institutions for laboratory diagnosis of fungal infections in addition to antifungal susceptibility tests that assist physicians to select the suitable treatment for each case.
Acknowledgments: None declared.
Ethical Permissions: None declared.
Conflicts of Interests: None declared.
Authors’ Contributions: Obeed FK (First Author), Introduction Writer/Main Researcher/Discussion Writer (34%); Al-Saad LA (Second Author), Assistant Researcher (33%); Alrubayae IMN (Third Author), Methodologist/Statistical Analyst (33%)
Funding/Support: None declared.
A hospital-acquired infection (HAI), also known as a nosocomial infection, is a form of infection that occurs in hospitals. The World Health Organization (WHO) describes HAIs as infections obtained while the patient is hospitalized but not present or incubating at the time of admission. The infections that occur more than 48 hours after admission to the hospitals are of big concern in the healthcare system, they are a leading cause of preventable disease. It has detrimental implications on both medical conditions and institutional efficiencies, including longer hospital visits, long-term injury, increased microorganism tolerance, substantial financial pressure (both patient and institution), and excess mortality [1, 2].
Nosocomial fungal infections are amongst the main causes of mortality in patients admitted to healthcare settings, especially in immunocompromised populations. The predominant pathogens include Candida spp., Aspergillus spp., Mucorales spp., and Fusarium spp. [3].
Over the last decades, the prevalence of invasive fungal infection has gradually increased, though, with a recent medical and surgical advancements, interventions, and an ever-growing population, the diversity and immunocompromised patients, also the number of fungal pathogens continues to increase, like Candida spp. are opportunistically invasive in people whose immune systems are weak [4, 5].
The risk and severity of infection increase in some special units in hospitals, such as intensive care units, child care units, burns, and units for patients with weak immunity [2].
Candida spp., especially C. albicans, are part of the human microbial flora; hence, most Candida infections are endogenous in origin. Candidemia disseminated hematogenous infections, or deep-seated infections can occur in immunocompromised patients, such as those with neutropenia, and critically ill patients [6].
In patients with chemotherapy-induced neutropenia and mucositis, candidemia may originate from the gastrointestinal tract. However, in critically ill patients, the source of candidemia is most likely a central vascular catheter (CVC) colonized by Candida spp. from the patient’s endogenous microflora or acquired from the health care environment [7].
Candida spp. have been isolated from environmental cultures of the floor, countertops, and other inanimate surfaces in the hospital. Patient acquisition and colonization with Candida spp. found in the hospital environment and food has been demonstrated. The propensity of Candida spp., especially C. parapsilosis, to cause Central line-associated bloodstream infections (CLABSIs) is likely related to this pathogen’s ability to form biofilms on catheters [3, 7].
Nosocomial infections are most difficult to diagnose, so early empiric therapy for patients who are clinically suspected of having a fungal infection, or prophylaxis for the highest-risk individuals, has been prioritized [4].
According to the scarcity of studies about nosocomial fungal infection and hospital community fungal profiling in Iraq, the current study was aimed at the prevalence of Hospital-Acquired Fungal Infections in Some Hospitals in Basrah Province.
Material and Methods
Two hundred thirteen clinical specimens were collected from patients after 48h of admission in Al–Faehaa Educational Hospital, AL-Sader Educational Hospital, and Pediatric Specialist Educational Hospital from November 2020 to April 2021, the age of patients ranged from 5 months to 80 years, 94 (44%) males and 119 (56%) females. The clinical specimens were sorted into 59 samples of midstream urine, 56 samples of wound swab, 29 samples of sputum, 27 samples of pleural fluid, 25 samples of blood, 13 samples of fluid, and 4 samples of pus. All clinical specimens were examined microscopically using the 10% KOH and Gram stain technique to show yeast cells and /or pseudohyphae or true hyphae and cultured on SDA with chloramphenicol and blood agar plates, the cultures were incubated at 37 °C for 1-14 days under aerobic conditions.
Yeast isolates were activated on SDA for 1-3 days to evaluate their ability of germ tube formation by adding a small portion of the activated colony to 0.5ml of blood serum. After that, the tubes were incubated at 37°C for 3 hours. The samples were investigated after the incubation period by adding a drop from inoculum on a clean slide and covered with a coverslip, then examined microscopically on 10X and 40X to detect the formation of the elongated tube without constriction from mother cells that indicated germ tube formation [8, 9].
Activated yeast isolates (1-3 days on SDA) were grown on Brilliance TM Candida Agar and incubated at 30°C for 1-3 days to be identified according to their colors and appearance on this test medium according to the following Table 1.
All yeast isolates were diagnosed by Vitek®2 system 7.01 and 8.01 software (BIOMERIEUX, USA) according to the manufacturer's instructions.
A molecular study was performed to confirm the identity of the clinical yeast isolates according to Mirhendi et al. [10]. Genomic DNA was extracted according to the instructions provided by the manufacturer of Presto Mini gDNA yeast kit (Geneaid, Taiwan) after cultivation of a single colony on SDA for 48h at 25ᵒC. The large subunit 28S region of rDNA was considered to identify the isolated fungi using the universal primers Null1 F-5’- GCA TAT CAA TAA GCG GAG GAA AAG -3’ and Null4 R-5’- GGT CCG TGT TTC AAG ACG G -3’. The PCR mixture was prepared in a total volume of 25μl contained 12.5μl Master Mix (Bioneer, Korea), 1.5μl of each primer,1.0μl of genomic DNA as a template, and 9μl of nuclease-free water. The reaction was performed using (Applied Bio System, USA) thermal cycler and the PCR conditions were 95oC for 1 min followed by 35 cycles of 95ᵒC for 1 min, 56ᵒC for 45 sec, and 72ᵒC for 1 min followed by 1 min of final extension at 72ᵒC. The PCR product was visualized on 2% agarose gel electrophoresis, the gel was pre-stained with ethidium bromide. The PCR product (20μl) of each sample was sent to Macrogen Company (Korea) for purification and sequencing, the sequences were processed by Chromas 2.6.6 and then identified using Basic Local Alignment Search Tool (BLAST) for multiple comparisons with deposited reference strains of National Center of Biotechnology Information (NCBI).
All statistical analysis was performed using Statistical Package for the Social Sciences (SPSS) version 24. The non-parametric variable was compared using the Chi-square for bivariate, while Kruskal-Wallis Test was implemented to analyze multiple comparisons of non-parametric variables.
Table 1) Expected reaction on Brilliance TM Candida Agar

Findings
The results showed that nosocomial fungi were isolated from 27.7% of the total collected specimens. The heights prevalence of fungal infections according to age group was recorded in 21-30 age group which is significantly exceeded the other groups (p<0.05), as well as the result was explained that female shown high prevalence (57.62%) of fungal infections, which significantly differed than males (p<0.05; Table 2).
Figure 1 illustrated that the diabetic patients tended to be significantly (p<0.05) more susceptible to fungal infections than other patients (43.7%), while 30% of the malignant patients were infected.
A total of 59 yeast species belonging to two genera were identified from HAFIs after the incubation period. C. albicans was recorded as the most common species 47.56%, followed by C. glabrata at 15.25%, while Naganishia globose appeared in one blood specimen at 1.6% (Table 3).
Table 2) Prevalence of nosocomial fungal infections according to age groups and sex


Figure 1) The fungal infections prevalence according to the presence of chronic diseases
The microscopic examination (Figure 2) determined that nosocomial fungal infection was concentrated in the midstream urine specimens (50%) which surpassed both pus and wound significantly (p=0.034) with no significant differences among all other specimens comparison.

Figure 2) The percentage of positive specimens by microscopic direct examination
The microscopic direct examination for clinical specimens Figure 3 (A) represented an aggregation of budding yeast cells in the sputum of a 56 years female patient who spent 3 months in the hospital for coronavirus medication, she had hypoxemia symptoms, so she was transferred to internal medicine ward, Whereas Figure 3 (B) was determined the appearance of true hyphae in wound swab gotten from female post-cesarean surgery patient.

Figure 3) Microscopic direct examination (40X): A- dry film (gram stain) of sputum appears yeasts body; B- wet film (10KOH) of wound swab showing the true hyphae
A total of 59 yeast species belonging to two genera were identified from HAFIs after the incubation period (Table 3). C. albicans (47.56%) was exceeded all other species significantly (p<0.05), followed by Candida glabrata (15.25%) then C. tropicalis with significant differences between them (p=0.05), while the less prevalence recorded in C. lusitaniea (3.39%) and N. globose (1.6%) respectively with no significant differences among them (p=0.157).
Table 3) Prevalence of clinical yeast isolation from HAFIs

Thirty-one (50%) yeast isolates exhibited the ability to generate germ tubes, as shown in Figure 4 the germ tube was determined as free of constriction tube from the mother cell, which is distinguished C. albicans and related species from other Candida spp. as all of these isolates were presumptively identified as C. albicans or C. dubliniensis.

Figure 4) Germ tube formation of C. albicans after 3h incubation at 37oC in human serum (40x)
The differential brilliance Candida agar results (Figure 5) revealed that 59 isolates were returned to the genus Candida: C. albicans, C. dubliniensis, C. glabrata, C. lusitaniea, C. parapsilosis, C. tropicalis, and C. krusei.

Figure 5) Identification of Candida spp. according to the color of colonies on Brilliance Candida agar 1,2,3,4 C. albicans; C. dubliniensis (green color); 5,6 C. glabrata, C. parapsilosis; C. lusitaniea (brown/ beige); 7 C. tropicalis (dark blue).
The results of the Vitek diagnosis (Table 4) showed that C. albicans was the high prevalent species (47.5%), which was significantly surpass the other species (p<0.05) followed by Stephanoascus ciforrii (21.5%), C. tropicalis (15.7%), C. glabrata (7.8%), C. parapsilosis (5.9%), while the lowest prevalence was recorded in C. famata, Trichosporon asahi, C. lusitaniea (3.9%), and C. dubliniensis (2%) with no significant differences (p=0.157).
Table 4) The prevalence of isolated fungi according to the Vitek system diagnosis

Molecular diagnosis confirmed the identity of the yeast isolates. The sequence analysis of the large subunit of the rDNA (LSU) region that 37 isolates were identified as a Candida spp., which were deposited in the national center for biotechnology information (NCBI) under the accession numbers listed in Table 5.
Table 5) New recording isolates in gen bank

On the other hand, one yeast isolate was identified as N. globose A56_NL which was documented as a new record for the first time in Iraq and deposited in NCBI under the accession number OK597185. Relatively, the new recorded N. globose was isolated from the blood specimens of a 72-years old female patient lying in the burn unit who died after one week of her admission. The heavy growth appeared on blood agar and SDA after 4 days of incubation at 37⁰C as white, dry, irregular colonies, the microscopic examination showed that the yeast cells had broad true hyphae (Figure 6).

Figure 6) Microscopic examination of N. globose showed that yeast cells and true hyphae (40X)
Furthermore, the results of molecular identification (Table 6) revealed that 98.4% of yeast isolates were identified as a Candida spp., among them C. albicans (45.9%) had the superiority over the other species with a significant difference (p<0.05) followed by C. glabrata (14.7%), while C. lusitaniea, N. globose, and N. diffluens recorded the lowest prevalence with no significant differences among them (p=0.157).
Table 6) The prevalence of yeast isolates due to molecular diagnosis

The neighbor-joining tree represented the clustering of the tested isolates according to the related species, which is reflects the sequence homogeneity of the related individuals and the sensitivity of the 28S region of diagnose the fungi to the species level efficiently as it seems that this region is polymorphic enough to distinguish among species and could consider as a good marker for fungi molecular identification. Alternatively, three isolates deviated from this settlement, namely A13, A18, and A30 belonging to three different species that include C. albicans, C. tropicalis, and C. parapsilosis respectively (Figure 7). Also, it illustrated the relationship between one-way sequences (non-assembled, forward, or reverse only) for the same region (28S), and the results revealed the grouping of C. albicans isolates (A4, A11, A12, A14, A15, A17, A20, A27) in one cluster, whilst, the isolates A8 and A15 of C. parapsilosis and C. tropicalis, as well as isolates A7, A16, and A9 of C. albicans and C. parapsilosis were isolated in independent clusters. The cluster pattern here is similar to that of assembled sequences with less accuracy as the one-way sequence mostly being shorter with more noise on both edges, which leads to losing valuable pieces of polymorphic sectors (Figure 8). The results of both trees may recommend considering the assembled sequence for the 28S marker as it seemed that the polymorphic area of that region is relatively narrow.

Figure 7) Neighbor-joining tree of the 28S region of assembled sequences. The numbers represent the genetic distance among the isolates

Figure 8) Neighbor-joining tree of Non-Assembled sequences of the 28S region. The numbers represent the genetic distance among the isolates
The results of the molecular method were relatively similar to brilliance candida agar, while the Vitek diagnosis showed inconsistency in 30% of results (Table 7) because the vitek diagnosis depends on the stored database of it that requires updating each six months interval.
Table 7) Comparison among brilliance test, VitekR2 assay, and molecular method

Discussion
The diabetic patients were tend to be significantly (p<0.05) more susceptible to fungal infections than other patients (43.7%), while 30% of the malignant patients were infected, this result was agreement with Al-Duboon [11] and Alrubayae et al. [12]. This is suggests that the high blood sugar can decrease its supplement to the cells which leads to cell weakness of the phagocytes that triggers the virulence of opportunistic microorganisms, including fungi.
The total of 59 yeast species belonging to two genera were identified from HAFIs after incubation
period. C. albicans was recorded as the most common species 47.56%, followed by C. glabrata 15.25%, while N. globose was appeared in one blood specimen with 1.6%. The finding of current study was agreed with other researches that showed C. albicans as the first fungal causative agent caused opportunistic infections in immunocompromised patients [12-15].
The total of 59 yeast species belonging to two genera were identified from HAFIs after incubation period. C. albicans (47.56%) was exceeded all other species, followed by C. glabrata (15.25%) then C. tropicalis, while the less prevalence recorded in C. lusitaniea (3.39%) and N. globose (1.6%) respectively. Similarly, previous studies showed that C. albicans was the most common fungal agent that caused opportunistic infections in immunocompromised patients [12-15]. The ability of C. albicans to affect divers host functions is indeed could be associated with wide range of pathogenic factors and physiological characteristics such as immediate adaptation to pH variation, metabolic adaptability, powerful nutrient acquisition systems and vigorous stress response machineries [16].
Thirty one (50%) yeast isolates exhibited the ability to generate germ tubes. The germ tube was determined as free of constriction tube from the mother cell, which is distinguished C. albicans and related species from other Candida spp. as all of these isolates were presumptively identified as C. albicans or C. dubliniensis. It has become clear that the physiological conditions of immunocompromised patients motivate the dimorphism in C. albicans from budding yeast to hyphal form, therefore, the growth conditions are induced in the laboratory whereby C. albicans grows as yeast only or produce the elongated tube (hyphal state) [17]. These result agreed with Alrubayae et al. [12] and Abbas et al. [15] who reported that almost tested isolates has the ability to form germ tube, which plays an important role of pathogenicity by assistants the pathogen to penetrate host cells barriers for nutrition acquisition [18]. Furthermore, the ability of most isolates to produce other virulence factors that increased their pathogenicity and facilitate penetration and adherence of host tissues like secreted hydrolytic enzymes (proteinase, phospholipase and hemolysis) and biofilm production [19].
The differential brilliance Candida agar results revealed that 59 isolates were returned to the genus Candida. The differential brilliance Candida agar medium mostly considered as primary assay to diagnose and could be differentiate among Candida species [10, 12, 13]. The results of molecular method were relatively similar to brilliance candida agar, while the Vitek diagnosis showed inconsistency in 30% of results, because the vitek diagnoses depends on the stored database of it that requires updating each six months interval, this is agreed with a previous findings performed by Abu-Mejdad et al. [20] and Alrubayae et al. [12].
Conclusion
The current study showed that fungi have an important role as etiological agents of hospital-acquired infections; so, it needs more attention from health institutions for laboratory diagnosis of fungal infections in addition to antifungal susceptibility tests that assist physicians to select the suitable treatment for each case.
Acknowledgments: None declared.
Ethical Permissions: None declared.
Conflicts of Interests: None declared.
Authors’ Contributions: Obeed FK (First Author), Introduction Writer/Main Researcher/Discussion Writer (34%); Al-Saad LA (Second Author), Assistant Researcher (33%); Alrubayae IMN (Third Author), Methodologist/Statistical Analyst (33%)
Funding/Support: None declared.
Article Type: Original Research |
Subject:
Health Promotion Setting
Received: 2022/01/28 | Accepted: 2022/04/12 | Published: 2022/04/30
Received: 2022/01/28 | Accepted: 2022/04/12 | Published: 2022/04/30
References
1. Allegranzi B, Boyce J, Chraiti M, Larson E, Pittet D, Sax H. The World Health Organization hand hygiene observation method. Am J Infect Control. 2009;37(10):827-34. [Link] [DOI:10.1016/j.ajic.2009.07.003]
2. Magill SS, O'Leary E, Janelle SJ, Thompson DL, Dumyati G, Nadle, J et al. Emerging infections program hospital prevalence survey team: changes in prevalence of health care-associated infections in US hospitals. N Engl J Med. 2018;379(18):1732-44. [Link] [DOI:10.1056/NEJMoa1801550]
3. Suleyman G, Alangaden GJ. Nosocomial fungal infections: epidemiology, infection control, and prevention. Infect Dis Clin North Am. 2016;30(4):1023-52. [Link] [DOI:10.1016/j.idc.2016.07.008]
4. Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007;45(4):321-46. [Link] [DOI:10.1080/13693780701218689]
5. Jose NV, Mudhigeti N, Asir J, Chandrakesan SD. Detection of virulence factors and phenotypic characterization of Candida isolates from clinical specimens. J Curr Res Sci Med. 2015;1(1):27-31. [Link]
6. Sanjukta B, Dutta AK. A comparative study on isolation and distribution of candida species with special reference to the risk factors associated with candidiasis. Int J Cell Biol Cell Process. 2021;7:66-70. [Link]
7. Blankenheim Y, Salmanton‐García J, Seifert H, Cornely OA, Koehler P. Attributable mortality of candidemia at a German tertiary hospital from 1997 to 2001 before the introduction of echinocandins. Mycoses. 2022;65(2):211-21. [Link] [DOI:10.1111/myc.13406]
8. Forbes BE, Sahm DF, Weissfeld AS. Bailey and scotts diagnostic microbiology. 12 Edition. Texas: Elsevier; 2007. [Link]
9. Sagar Aryal S. Germ tube test-principle, procedure, results, interpretation and limitations [Internet]. MicrobiologyInfo; 2015 [Cited 2020 Jul 01?]. Available from: https://microbiologyinfo.com/germ-tube-test-principle-procedure-results-interpretation-and-limitations/ [Link]
10. Mirhendi SH, Adin H, Shidfar MR, Kordbacheh P, Hashemi SJ, Moazeni M et al. Identification of pathogenic Candida species: PCR-fragment size polymorphism (PCR-FSP) method. Tehran Univ Med J. 2008;66(9):639-45. [Persian] [Link]
11. Al-Duboon AH. Candiduria and urinary candidiasis in Basrah, Iraq. J Basrah Res Sci. 2010;36(1). [Link]
12. Alrubayae IM, Al-Duboon AH, Majed M. Study of Candida spp. associated with urinary tract infections in Basra: urinary tract infection associated with Candida spp: Their identification, pathogenicity and susceptibility to antifungals. Germany: LAP LAMBERT Academic Publishing; 2013. p. 196. [Link]
13. Allaaeiby AIE, Al-Mousawi AA, Alrubayae I, Al-Saadoon A, Almayahi M. Innate pathogenic traits in oral yeasts. Karbala Int J Modern Sci. 2020;6(4):5. [Link] [DOI:10.33640/2405-609X.1984]
14. Alrubayae IM, Al-laaeiby A, Minati MH, ALibraheem SA. Determination of genetic relationships and pathogenicity of oral candidiasis etiological agents in pediatric malignant patients in Basrah province, Iraq. Sys Rev Pharm. 2020;11. [Link]
15. Abbas NF, Shani WS, Alrubyae IM. Evaluation of immunization efficacy for cell wall fraction antigen separated from clinical isolate of Candida albicans. Technium Bio Chem Med. 2021;2(2):60-76. [Link]
16. Matare T, Nziramasanga P, Gwanzura L, Robertson V. Experimental germ tube induction in Candida albicans: An evaluation of the effect of sodium bicarbonate on morphogenesis and comparison with pooled human serum. Biomed Res Int. 2017;2017:1976273. [Link] [DOI:10.1155/2017/1976273]
17. Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4(2):119-28. [Link] [DOI:10.4161/viru.22913]
18. Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12(7):317-24. [Link] [DOI:10.1016/j.tim.2004.05.008]
19. Abeed FK, Alrubayae IM. Evaluation of virulence factors of clinical yeast isolates from nosocomial fungal infections with the determination of their antifungal susceptibility profile. Iran J Ichthyol. 2022;9:61-68. [Link]
20. Abu-Mejdad NM, Al-Badran AI, Al-Saadoon AH. A Novel report on killer yeast strains identification methods. Basrah J Agricult Sci. 2020;33(1):39-49. [Link] [DOI:10.37077/25200860.2020.32.1.04]
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |





