Volume 12, Issue 3 (2024)
Health Educ Health Promot 2024, 12(3): 537-545 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Mohammadi P, Noruzinia M, Ebadi M, Ghoraeian P. Identification of Blood Biomarkers for Veno-Occlusive Disease for Early Diagnosis and Enhancing Patients Quality of Life. Health Educ Health Promot 2024; 12 (3) :537-545
URL: http://hehp.modares.ac.ir/article-5-76901-en.html
URL: http://hehp.modares.ac.ir/article-5-76901-en.html
1- Department of Biology, Damghan Branch, Islamic Azad University, Damghan, Iran
2- Department of Medical Genetics, Faculty of Medicine, Tarbiat Modares University, Tehran, Iran
3- Department of Genetics, Faculty of Advanced Science and Technology, Tehran Medical Sciences, Islamic Azad University, Tehran, Iran
2- Department of Medical Genetics, Faculty of Medicine, Tarbiat Modares University, Tehran, Iran
3- Department of Genetics, Faculty of Advanced Science and Technology, Tehran Medical Sciences, Islamic Azad University, Tehran, Iran
Keywords: Pulmonary Veno-Occlusive Disease [MeSH], miRNA [MeSH], Biomarker [MeSH], Quality of Life [MeSH], Biomarkers [MeSH]
Full-Text [PDF 897 kb]
(1995 Downloads)
| Abstract (HTML) (729 Views)
Full-Text: (149 Views)
Introduction
Veno-occlusive disease (VOD), or sinusoidal obstruction syndrome (SOS), is a serious and potentially lethal condition most commonly observed in patients undergoing hematopoietic stem cell transplantation (HSCT) [1]. VOD/SOS manifests as hepatomegaly, right upper quadrant (RUQ) pain, jaundice, and ascites, resulting from sinusoidal endothelial cell injury and subsequent portal hypertension [2]. Accurate and timely diagnosis is crucial to enable early intervention and improve patient outcomes. Key factors in assessing risk include patient-specific and transplantation-related factors that can aid in stratifying patients [3]. Management strategies for VOD/SOS encompass supportive care, intensive monitoring, and specific drug therapies such as defibrotide, which is commonly used to manage hepatic VOD and thrombotic thrombocytopenic purpura (TTP) [4]. Antimicrobial prophylaxis remains under study, and further research is ongoing to address resolution strategies for high-risk patients [5]. A recent decline in mortality rates has been observed, likely due to more intensive, multidisciplinary approaches to managing VOD/SOS.
Despite advancements in HSCT procedures and supportive care, the diagnosis and effective management of VOD remain challenging due to the limitations of current diagnostic methods. Several prior studies have focused on identifying less invasive biomarkers for diagnosing VOD and liver fibrosis. Cutler et al. found that biomarkers of endothelial damage, particularly von Willebrand Factor and thrombomodulin, demonstrate high sensitivity and specificity in identifying VOD in patients receiving sirolimus [6]. Imaging techniques, such as ultrasound with Doppler evaluation and elastography, are helpful in diagnosing VOD, although their reliability continues to be evaluated. In non-alcoholic fatty liver disease (NAFLD), serum markers related to inflammation, apoptosis, and oxidative stress have been studied in depth [7]. Transient elastography and other imaging techniques are increasingly used in managing fibrosis across various chronic liver diseases. The authors emphasize the need to investigate non-invasive markers for liver fibrosis assessment, given the risks and limitations associated with liver biopsy [8].
Recent advances in bioinformatics and molecular biology have created new opportunities to identify potential blood biomarkers associated with VOD. High-throughput sequencing technologies, such as next-generation sequencing (NGS) and mass spectrometry, have enabled unprecedented large-scale analyses of genetic and proteomic data [9]. These technologies allow for the simultaneous examination of thousands of genes and proteins. Bioinformatics techniques are crucial for analyzing this large-scale data, as they help identify novel biomarkers by uncovering patterns, classifying candidates, and revealing gene interactions. Despite its pivotal role in biomarker discovery and development, bioinformatics faces several limitations. For example, the absence of efficient computational strategies can impede discovery, highlighting the need for innovative approaches, such as graph-based scoring functions, to improve candidate ranking in proteomic data analysis [10]. Additionally, enhanced database linkages and high-quality input data are essential for advancing biomarker development [11].
Given recent advancements and ongoing challenges, the identification of blood biomarkers could significantly enhance the treatment of VOD, especially considering the limited understanding of the disease’s underlying mechanisms. Identifying these biomarkers could lead to improvements in both diagnostic and therapeutic strategies, ultimately enhancing patient quality of life and outcomes. Blood biomarkers could serve as diagnostic markers for early disease detection, which is often challenging since clinical manifestations appear at advanced stages when organ damage may be irreversible. Early detection of VOD is crucial, as patients treated in the initial stages tend to respond more effectively to therapy, with prognosis reportedly improving significantly with early intervention. Additionally, these biomarkers could aid in monitoring disease progression and patient response to prescribed medications, supporting the development of therapeutic strategies that are less toxic yet more effective. This article introduced a set of potential blood biomarkers for VOD identified through bioinformatics analysis, with the goal of improving diagnostic methods and informing therapeutic strategies for managing high-risk patients and those with severe VOD. This study aimed to identify novel blood biomarkers associated with VOD through bioinformatics analysis to improve early diagnosis and treatment outcomes.
Materials and Methods
Data collection and sample grouping
This retrospective cohort study, which was conducted at the Islamic Azad University, Damghan Branch in 2024, employed GSE164635 for dataset selection [12], comprising a total of 30 samples. These samples were categorized into three distinct groups, including a control group with nine samples, a group of nine samples from patients diagnosed with VOD, and a third group with 12 samples from individuals with other forms of liver disease. This systematic grouping enabled a comparative analysis aimed at identifying novel miRNA biomarkers associated with the early diagnosis of VOD. To ensure the dataset’s quality and reliability, we performed data validation steps, including statistical checks for inconsistencies and outliers. We assessed the expression level distributions and conducted statistical tests to confirm the absence of significant outliers, ensuring the dataset’s robustness for further analysis.
Data analysis and miRNA selection
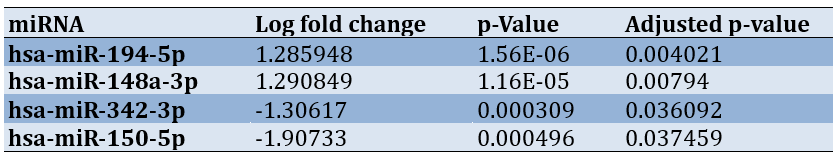
Data analysis was conducted using R software. Initially, the raw expression data were normalized using the Affy package, which employs robust multi-array averaging (RMA) to adjust for background noise and normalize data across samples. This step ensures the comparability of expression levels and reduces technical variation. Following normalization, differential expression analysis was performed with the Limma package [13]. A log fold change (logFC) filter was applied, setting criteria at greater than +1 or less than -1, along with an adjusted p-value (adj.P.Val) of less than 0.05. These thresholds were chosen to identify biologically significant changes in miRNA expression while minimizing false positives. A logFC threshold of ±1 corresponds to at least a doubling or halving of expression, which is biologically meaningful. The adj.P.Val threshold of 0.05 is commonly used in genomic studies to control for multiple testing errors, ensuring statistical significance for the identified miRNAs. Using these stringent filtering criteria, we identified four significant miRNAs, namely two overexpressed (hsa-miR-194-5p and hsa-miR-148a-3p) and two underexpressed (hsa-miR-342-3p and hsa-miR-150-5p) miRNAs (Table 1).
Table 1. Dysregulated miRNAs
A volcano plot was generated to visually depict the distribution of these miRNAs based on their logFC and adjusted p-values.
miRNA target prediction
Gene targets for the identified miRNAs were predicted using the multiMiR package [14], which draws data from the mirTarBase and Tarbase databases. A Venn diagram was created to identify common gene targets for the overexpressed miRNAs (hsa-miR-194-5p and hsa-miR-148a-3p), revealing 159 genes shared between the two databases. Similarly, a Venn diagram for the underexpressed miRNAs (hsa-miR-342-3p and hsa-miR-150-5p) identified 74 shared gene targets.
Pathway enrichment and gene ontology analysis
Pathway enrichment and gene ontology (GO) analyses were conducted on the shared genes using Funrich 3.1.3 software. These analyses are essential for understanding the biological significance and functional roles of the genes identified in VOD. The analyses were grouped into four categories, including biological processes (BPs), cellular components, molecular functions (MFs), and biological pathways.
The BP category encompassed a range of cellular and organismal activities critical to function. Analyzing BPs allowed us to identify processes significantly associated with shared genes, providing insights into their roles in VOD development and progression. Notable processes included inflammatory responses, cell adhesion, apoptosis, and various metabolic pathways.
The cellular component category identified the cellular locations where gene products (proteins) are active, enhancing our understanding of the subcellular structures implicated in VOD pathology. Key cellular components include the plasma membrane, nucleus, cytoplasm, and extracellular matrix. This analysis helps localize critical molecular interactions within VOD-affected cells.
The MF category described the specific biochemical activities of gene products, such as enzymatic functions, binding affinity, and signal transduction. Examining MFs enabled us to elucidate the primary functional roles of shared genes at a molecular level.
Network construction and Hub gene identification
Networks of common genes were constructed using Cytoscape software. The CytoHubba plugin [15] was employed to identify hub genes within these networks. A network comprising 87 nodes and 410 edges was constructed for the 159 genes associated with the overexpressed miRNAs, enabling the identification of potential key players in the pathogenesis of VOD.
Findings
Identification of differentially expressed miRNAs in VOD samples and target gene prediction
Using the GSE164635 dataset, which includes 30 samples (nine control samples, nine VOD patient samples, and 12 samples with other liver diseases), we identified four differentially expressed miRNAs (DEmiRNAs) through stringent filtration criteria (logFC > +1 or < -1 and adj.P.Val < 0.05). The identified DEmiRNAs were hsa-miR-194-5p, hsa-miR-148a-3p, hsa-miR-342-3p, and hsa-miR-150-5p. To validate our findings, we confirmed the association of these miRNAs with relevant diseases using the Human MicroRNA Disease Database (HMDD) [16], supporting the potential clinical relevance of these miRNAs in VOD. Notably, hsa-miR-194-5p was highlighted among the identified miRNAs, further underscoring its significance (Figure 1A).
Target gene prediction for these miRNAs was performed using the multiMiR package in R, with data sourced from the mirTarBase and Tarbase databases. For the two upregulated miRNAs (hsa-miR-194-5p and hsa-miR-148a-3p), 159 common target genes were identified. Similarly, 74 common target genes were identified for the two downregulated miRNAs (hsa-miR-342-3p and hsa-miR-150-5p). The overlap of the target genes for these miRNAs was depicted in Venn diagrams (Figure 1B and 1C).
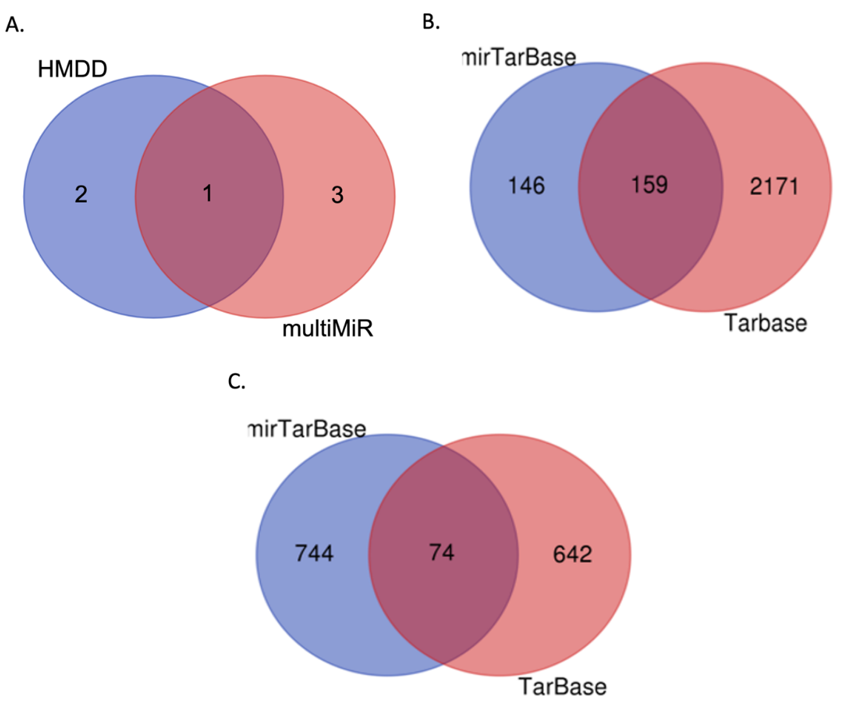
Figure 1. Venn diagram illustrating the shared miRNAs and predicted target genes.
A. Intersection of the four identified differentially expressed miRNAs (DEmiRNAs) and their confirmed associations with relevant diseases from the Human miRNA Disease Database (HMDD). B. Overlap of target genes predicted for the upregulated miRNAs (hsa-miR-194-5p and hsa-miR-148a-3p), showing 159 common targets. C. Overlap of target genes for the downregulated miRNAs (hsa-miR-342-3p and hsa-miR-150-5p), indicating 74 common targets
Gene ontology and pathway analysis of identified target genes
For the 159 target genes of the upregulated miRNAs (hsa-miR-194-5p and hsa-miR-148a-3p), pathway enrichment and GO analyses were conducted using Funrich 3.1.3 software. The identified target genes were significantly enriched in several biological pathways, including the VEG and VEGFR signaling network, plasma membrane estrogen receptor signaling, glypican 1 network, proteoglycan syndecan-mediated signaling events, glypican pathway, and integrin family cell surface interactions (Figure 2A). In terms of BPs, the enriched categories included gene silencing, morphogenesis, RNA metabolism, epigenetic regulation of gene expression, cell communication, and signal transduction (Figure 2B). Cellular component analysis revealed significant enrichment in the RNA-induced silencing complex, cytosol, micro-ribonucleoprotein complex, lysosomes, exosomes, and the cytoplasm (Figure 2C). The enriched terms for MFs included GTPase activity, receptor signaling protein serine/threonine kinase activity, MHC class II receptor activity, MHC class I receptor activity, protein tyrosine phosphatase activity, and DNA methyltransferase activity (Figure 2D).
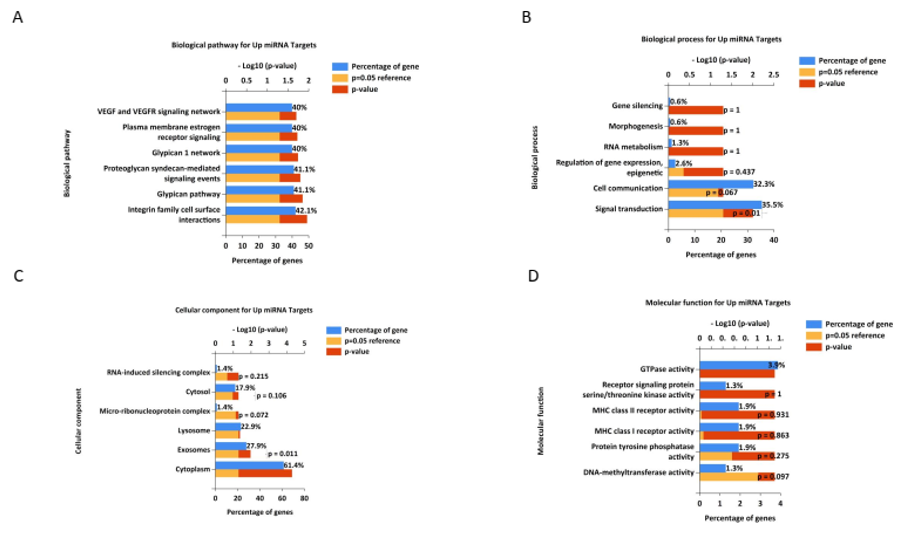
Figure 2. Gene ontology and pathway analysis of identified target genes for upregulated miRNAs. The most significant terms for biological process (BP), cellular component (CC), molecular function (MF), and biological pathway (Bipath) were identified and visualized using R software. Terms with a p-value and adjusted p-value<0.05 were considered significant
Gene-miRNA interaction for downregulated miRNAs
Similarly, for the 74 target genes of the downregulated miRNAs (hsa-miR-342-3p and hsa-miR-150-5p), pathway enrichment and GO analyses provided significant insights into the molecular mechanisms affected by these miRNAs. The biological pathways enriched for these target genes included validated transcriptional targets of AP1 family members Fra1 and Fra2, hypoxic and oxygen homeostasis regulation of HIF-1-alpha, the E2F transcription factor network, the HIF-2-alpha transcription factor network, the HIF-1-alpha transcription factor network, and the regulation of retinoblastoma proteins (Figure 3A). The BPs enriched in these target genes included transcription, lipid metabolism, protein folding, regulation of enzyme activity, vesicle docking, and regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolism (Figure 3B). Cellular component analysis revealed significant enrichment in the nodes of Ranvier, nuclear centromeric heterochromatin, protein complexes, membranes, endoplasmic reticulum membranes, and nuclei (Figure 3C). For molecular function, the enriched terms included receptor signaling protein tyrosine phosphatase activity, phosphatase regulator activity, kinase binding, DNA methyltransferase activity, protein tyrosine phosphatase activity, and transcription factor activity (Figure 3D).
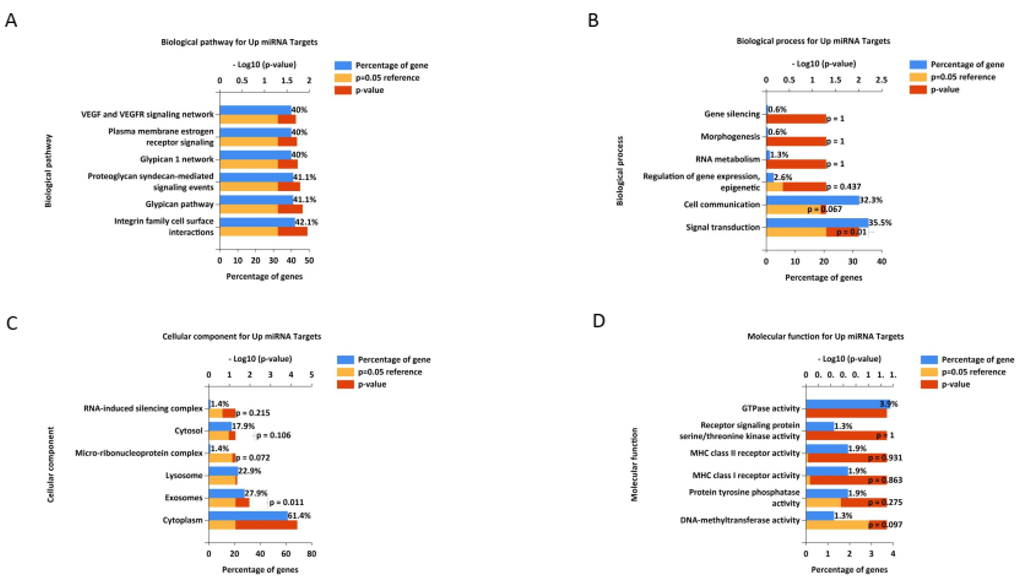
Figure 3. Gene ontology and pathway analysis of identified target genes for down-regulated miRNAs
Gene-miRNA interaction networks
Gene-miRNA interaction for up-regulated miRNAs: Gene-miRNA interaction networks for upregulated and downregulated miRNAs were constructed using Cytoscape. For the upregulated miRNAs, the network revealed interactions with key genes, such as AGO2, CDKN1A, HSP90AA1, HSPA4, EP300, IGF1R, MYC, SMAD2, DICER1, and IL10. This network (Figure 4A) highlighted the central roles of these genes in various cellular processes and pathways.
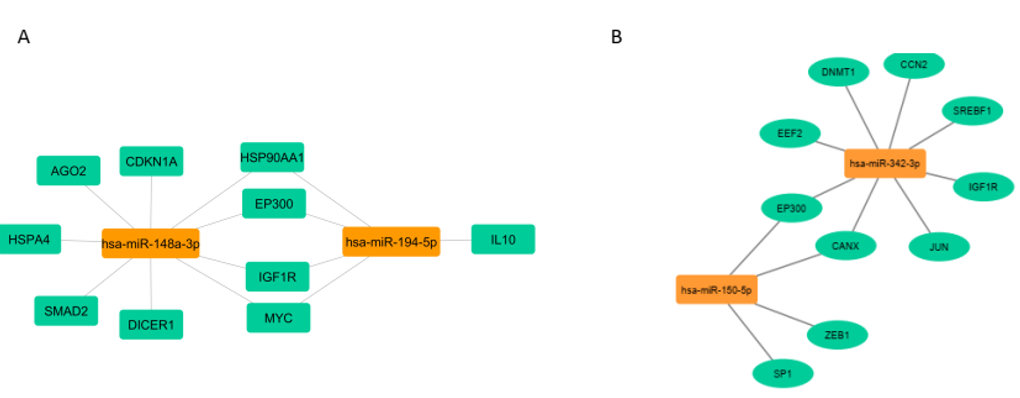
Figure 4. Visualization of miRNA-target gene interaction
Gene-miRNA interaction for down-regulated miRNAs: For the downregulated miRNAs, the interaction network identified significant genes, including EEF2, IGF1R, EP300, CCN2, DNMT1, SREBF1, CANX, ZEB1, SP1, and JUN. This network (Figure 4B) underscores the involvement of these genes in transcriptional regulation, lipid metabolism, protein folding, and other functions (Figure 4B).
These comprehensive analyses provide valuable insights into the molecular mechanisms underlying VOD and suggest potential biomarkers and therapeutic targets for improving the diagnosis and treatment of this condition.
Discussion
This study aimed to identify novel blood biomarkers associated with veno-occlusive disease through bioinformatics analysis to improve early diagnosis and treatment outcomes. VOD is a severe condition characterized by the blockage of small hepatic veins. It primarily occurs after chemotherapy and radiation, particularly following HSCT. VOD is associated with various causes, including obesity and underlying diseases such as sickle cell disease, certain leukemias, and other hematological disorders. The functions of microRNAs (miRNAs) are highly relevant in numerous physiological and pathological processes, including inflammation, liver fibrosis, and cellular stress responses, all of which are crucial in the development and progression of VOD. In this session, we will discuss the miRNA/gene pathways involved in the mechanisms of action related to VOD.
Our results demonstrated interactions between the upregulated miRNAs (hsa-miR-194-5p and hsa-miR-148a-3p) and key genes, including AGO2, CDKN1A, HSP90AA1, HSPA4, EP300, IGF1R, MYC, SMAD2, DICER1, and IL10 in VOD. A recent study found that patients and rats with pyrrolizidine alkaloid-induced hepatic sinusoidal obstruction syndrome (HSOS) exhibit overexpression of three miRNAs: miR-148a-3p, miR-362-5p, and miR-194-5p [12]. AGO2, a key component of the RNA-induced silencing complex (RISC), along with DICER1, which is an essential enzyme for the production of microRNAs (miRNAs), plays a critical role in regulating gene expression [17].
The insulin-like growth factor 1 receptor (IGF1R) is a transmembrane receptor that is crucial for growth and development. It mediates the effects of IGF-1, which is involved in various cellular processes, including proliferation, differentiation, and survival. IGF1R signaling is particularly important for liver function and has implications for various diseases, including cancer and metabolic disorders [18]. HSP90AA1, which encodes a molecular chaperone, and HSPA4, a heat shock protein, are essential in various cancers and diseases. HSP90AA1 promotes inflammation in the liver, potentially contributing to liver fibrosis [19, 20]. SMAD2 serves as a key mediator of the transforming growth factor beta (TGF-β) signaling pathway, which is critical in liver fibrosis. TGF-β is known to promote the activation of hepatic stellate cells (HSCs), leading to collagen deposition and fibrosis, which can exacerbate VOD [21, 22].
IL-10 is a potent anti-inflammatory cytokine that plays a protective role in various liver conditions, including VOD. It can inhibit the production of pro-inflammatory cytokines, thereby reducing inflammatory responses and tissue damage [23]. CDKN1A, also known as P21, is associated with cellular senescence and the cell cycle and is implicated in liver fibrosis and inflammation. Elevated CDKN1A expression has been observed in multiple cancer types after chemotherapy and radiotherapy [24].
EP300 (E1A-binding protein p300) is a transcriptional coactivator that plays a crucial role in regulating gene expression. It is involved in several cellular processes, including cell proliferation, differentiation, and apoptosis. EP300 is known for its histone acetyltransferase activity, which modifies chromatin and facilitates gene transcription by recruiting various transcription factors and RNA polymerase II to target genes [25]. Research suggests that EP300 inhibition can attenuate fibrosis-related molecular processes, including extracellular matrix deposition, inflammation, and epithelial-to-mesenchymal transition [26].
MYC is a family of regulatory genes that encode transcription factors involved in various cellular processes, such as cell proliferation, growth, differentiation, and apoptosis. MYC plays a significant role in oncogenesis and is dysregulated in various cancers. Aberrant MYC expression can lead to increased cell proliferation and resistance to apoptosis, contributing to tumor progression and metastasis [27].
Eukaryotic elongation factor 2 (EEF2) is a critical protein involved in protein synthesis (translation) within eukaryotic cells. It plays a vital role in the elongation phase of translation, facilitating the movement of the ribosome along the messenger RNA (mRNA) strand as amino acids are added to the growing polypeptide chain [28].
To date, no studies have suggested a connection between VOD and the expression of MYC, EEF2, or EP300. Nevertheless, these factors may influence pathways associated with VOD.
Furthermore, we established a network linking the downregulated hsa-miR-342-3p and hsa-miR-150-5p to their targets EEF2, IGF1R, EP300, CCN2, DNMT1, SREBF1, CANX, ZEB1, SP1, and JUN. In hepatocellular carcinoma (HCC), miR-342-3p acts as a potent tumor suppressor [29]. Additionally, miR-342-3p influences HCC cell proliferation and has potential as a target for investigating therapeutics [30].
Hsa-miR-150-5p is a microRNA (miRNA) that has been studied in relation to lipid metabolism. Various studies suggest that it may play a role in hepatic lipid homeostasis by influencing lipid accumulation in the liver [31]. Another study showed that miR-150-5p exhibits opposite regulation and function in HSCs and hepatocytes during liver fibrosis [32].
Cellular communication network 2 (CCN2) is involved in cell migration, angiogenesis, and tissue repair. Studies have reported that CCN2 is associated with various aspects of lipid metabolism, including adipogenesis, lipolysis, lipid synthesis, and lipid oxidative stress [33, 34].
DNMT1, or DNA methyltransferase 1, is involved in DNA methylation, a key epigenetic mechanism. Nutrient-rich diets may upregulate DNA methyltransferases, leading to hypermethylation of the Klb promoter and downregulation of Klb expression, which impairs fatty acid oxidation and contributes to the development of NAFLD in male C57BL/6J mice [35].
SREBF1, also known as sterol regulatory element-binding transcription factor 1, regulates genes involved in lipid metabolism. SREBPs can participate in the development of liver fibrosis through a lipid-independent pathway. However, SREBP-1c can inhibit the activation of HSCs and liver fibrosis by downregulating TGF-β, SMAD3, and AKT [36].
Calnexin (CANX) is a protein involved in protein folding in the endoplasmic reticulum. Recent studies suggest that calnexin does more than assist with protein folding; it also interacts with other proteins involved in signaling and lipid metabolism. One of these proteins, PTP-1B, is attached to the ER membrane, similar to DGAT2. The connection between PTP-1B and the ER relies on calnexin [37].
ZEB1, or zinc finger E-box binding homeobox 1, is a transcription factor involved in cell differentiation and epithelial-mesenchymal transition. SP1, or Specificity Protein 1, is a transcription factor implicated in various BPs, including cell growth and apoptosis. Research has shown that miR-124 targets specificity protein 1 (Sp1), an important gene for liver lipid metabolism [38, 39].
JUN is a transcription factor involved in cell growth, apoptosis, and differentiation. Gao et al. reported that in the presence of non-esterified fatty acids (NEFA), activation of the inositol-requiring enzyme 1α and c-Jun N-terminal kinase axis contributes to intracellular lipid accumulation in calf hepatocytes [40].
Continuous research is essential for further exploration of the roles of miRNAs in VOD and for uncovering their potential clinical applications. This study highlights the significant role of miRNAs in VOD, suggesting potential biomarkers for early diagnosis and intervention. Identifying these biomarkers could lead to improved screening methods for high-risk populations, allowing for timely therapeutic strategies that may mitigate the severity of VOD. Moreover, understanding the molecular mechanisms underlying VOD can inform preventive measures, particularly in patients undergoing HSCT or those with predisposing conditions. By targeting specific miRNA pathways, future therapies could not only treat existing conditions but also prevent the onset of VOD in susceptible individuals. Further investigation is needed to identify specific miRNAs. Animal models are being employed to examine the impact of specific miRNAs on the pathophysiology of the disease. Additionally, advancements in genomics may pave the way for clinical studies that establish a correlation between miRNA profiles and VOD outcomes in patients. This field could potentially lead to the discovery of innovative biomarkers for diagnosis and the development of novel therapeutic approaches for managing VOD.
While our findings contribute to the understanding of miRNA pathways in VOD, several limitations should be considered. The study’s reliance on specific models may not fully represent the complexity of VOD in human patients. Additionally, the sample size for some miRNA analyses was limited, which could affect the generalizability of the findings. Further research with larger cohorts and diverse populations is necessary to validate these results and explore the clinical implications of the identified biomarkers.
Conclusion
The pathophysiology of VOD is greatly influenced by microRNAs, which play a crucial role in regulating inflammation, fibrosis, endothelial function, and cellular survival.
Acknowledgments: We are sincerely grateful for the support of the professors and staff of Islamic Azad University, Damghan Branch during the conduct of this research. We also acknowledge the contributions of the researchers who provided access to the GSE164635 dataset and the developers of the R packages used in our analysis. Special thanks to our colleagues for their valuable views and feedback during the study
Ethical Permissions: This study was conducted in accordance with the ethical standards of Islamic Azad University, Damghan branch. Ethical approval was obtained from the university's Institutional Review Board (IRB). All patient data were anonymized to ensure confidentiality and conform to ethical guidelines for research involving human subjects.
Conflicts of Interests: The authors reported no conflicts of interests.
Authors' Contribution: Mohammadi P (First Author), Introduction Writer/Main Assistant Researcher/Discussion Writer (60%); Noruzinia M (Second Author), Methodologist/Original or Assistant Researcher/Discussion Writer/Statistical Analyst (20%); Ebadi M (Third Author), Assistant Researcher (10%); Ghoraeian P (Fourth Author), Discussion Writer/Statistical Analyst (10%)
Funding/Support: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Veno-occlusive disease (VOD), or sinusoidal obstruction syndrome (SOS), is a serious and potentially lethal condition most commonly observed in patients undergoing hematopoietic stem cell transplantation (HSCT) [1]. VOD/SOS manifests as hepatomegaly, right upper quadrant (RUQ) pain, jaundice, and ascites, resulting from sinusoidal endothelial cell injury and subsequent portal hypertension [2]. Accurate and timely diagnosis is crucial to enable early intervention and improve patient outcomes. Key factors in assessing risk include patient-specific and transplantation-related factors that can aid in stratifying patients [3]. Management strategies for VOD/SOS encompass supportive care, intensive monitoring, and specific drug therapies such as defibrotide, which is commonly used to manage hepatic VOD and thrombotic thrombocytopenic purpura (TTP) [4]. Antimicrobial prophylaxis remains under study, and further research is ongoing to address resolution strategies for high-risk patients [5]. A recent decline in mortality rates has been observed, likely due to more intensive, multidisciplinary approaches to managing VOD/SOS.
Despite advancements in HSCT procedures and supportive care, the diagnosis and effective management of VOD remain challenging due to the limitations of current diagnostic methods. Several prior studies have focused on identifying less invasive biomarkers for diagnosing VOD and liver fibrosis. Cutler et al. found that biomarkers of endothelial damage, particularly von Willebrand Factor and thrombomodulin, demonstrate high sensitivity and specificity in identifying VOD in patients receiving sirolimus [6]. Imaging techniques, such as ultrasound with Doppler evaluation and elastography, are helpful in diagnosing VOD, although their reliability continues to be evaluated. In non-alcoholic fatty liver disease (NAFLD), serum markers related to inflammation, apoptosis, and oxidative stress have been studied in depth [7]. Transient elastography and other imaging techniques are increasingly used in managing fibrosis across various chronic liver diseases. The authors emphasize the need to investigate non-invasive markers for liver fibrosis assessment, given the risks and limitations associated with liver biopsy [8].
Recent advances in bioinformatics and molecular biology have created new opportunities to identify potential blood biomarkers associated with VOD. High-throughput sequencing technologies, such as next-generation sequencing (NGS) and mass spectrometry, have enabled unprecedented large-scale analyses of genetic and proteomic data [9]. These technologies allow for the simultaneous examination of thousands of genes and proteins. Bioinformatics techniques are crucial for analyzing this large-scale data, as they help identify novel biomarkers by uncovering patterns, classifying candidates, and revealing gene interactions. Despite its pivotal role in biomarker discovery and development, bioinformatics faces several limitations. For example, the absence of efficient computational strategies can impede discovery, highlighting the need for innovative approaches, such as graph-based scoring functions, to improve candidate ranking in proteomic data analysis [10]. Additionally, enhanced database linkages and high-quality input data are essential for advancing biomarker development [11].
Given recent advancements and ongoing challenges, the identification of blood biomarkers could significantly enhance the treatment of VOD, especially considering the limited understanding of the disease’s underlying mechanisms. Identifying these biomarkers could lead to improvements in both diagnostic and therapeutic strategies, ultimately enhancing patient quality of life and outcomes. Blood biomarkers could serve as diagnostic markers for early disease detection, which is often challenging since clinical manifestations appear at advanced stages when organ damage may be irreversible. Early detection of VOD is crucial, as patients treated in the initial stages tend to respond more effectively to therapy, with prognosis reportedly improving significantly with early intervention. Additionally, these biomarkers could aid in monitoring disease progression and patient response to prescribed medications, supporting the development of therapeutic strategies that are less toxic yet more effective. This article introduced a set of potential blood biomarkers for VOD identified through bioinformatics analysis, with the goal of improving diagnostic methods and informing therapeutic strategies for managing high-risk patients and those with severe VOD. This study aimed to identify novel blood biomarkers associated with VOD through bioinformatics analysis to improve early diagnosis and treatment outcomes.
Materials and Methods
Data collection and sample grouping
This retrospective cohort study, which was conducted at the Islamic Azad University, Damghan Branch in 2024, employed GSE164635 for dataset selection [12], comprising a total of 30 samples. These samples were categorized into three distinct groups, including a control group with nine samples, a group of nine samples from patients diagnosed with VOD, and a third group with 12 samples from individuals with other forms of liver disease. This systematic grouping enabled a comparative analysis aimed at identifying novel miRNA biomarkers associated with the early diagnosis of VOD. To ensure the dataset’s quality and reliability, we performed data validation steps, including statistical checks for inconsistencies and outliers. We assessed the expression level distributions and conducted statistical tests to confirm the absence of significant outliers, ensuring the dataset’s robustness for further analysis.
Data analysis and miRNA selection
Data analysis was conducted using R software. Initially, the raw expression data were normalized using the Affy package, which employs robust multi-array averaging (RMA) to adjust for background noise and normalize data across samples. This step ensures the comparability of expression levels and reduces technical variation. Following normalization, differential expression analysis was performed with the Limma package [13]. A log fold change (logFC) filter was applied, setting criteria at greater than +1 or less than -1, along with an adjusted p-value (adj.P.Val) of less than 0.05. These thresholds were chosen to identify biologically significant changes in miRNA expression while minimizing false positives. A logFC threshold of ±1 corresponds to at least a doubling or halving of expression, which is biologically meaningful. The adj.P.Val threshold of 0.05 is commonly used in genomic studies to control for multiple testing errors, ensuring statistical significance for the identified miRNAs. Using these stringent filtering criteria, we identified four significant miRNAs, namely two overexpressed (hsa-miR-194-5p and hsa-miR-148a-3p) and two underexpressed (hsa-miR-342-3p and hsa-miR-150-5p) miRNAs (Table 1).
Table 1. Dysregulated miRNAs

A volcano plot was generated to visually depict the distribution of these miRNAs based on their logFC and adjusted p-values.
miRNA target prediction
Gene targets for the identified miRNAs were predicted using the multiMiR package [14], which draws data from the mirTarBase and Tarbase databases. A Venn diagram was created to identify common gene targets for the overexpressed miRNAs (hsa-miR-194-5p and hsa-miR-148a-3p), revealing 159 genes shared between the two databases. Similarly, a Venn diagram for the underexpressed miRNAs (hsa-miR-342-3p and hsa-miR-150-5p) identified 74 shared gene targets.
Pathway enrichment and gene ontology analysis
Pathway enrichment and gene ontology (GO) analyses were conducted on the shared genes using Funrich 3.1.3 software. These analyses are essential for understanding the biological significance and functional roles of the genes identified in VOD. The analyses were grouped into four categories, including biological processes (BPs), cellular components, molecular functions (MFs), and biological pathways.
The BP category encompassed a range of cellular and organismal activities critical to function. Analyzing BPs allowed us to identify processes significantly associated with shared genes, providing insights into their roles in VOD development and progression. Notable processes included inflammatory responses, cell adhesion, apoptosis, and various metabolic pathways.
The cellular component category identified the cellular locations where gene products (proteins) are active, enhancing our understanding of the subcellular structures implicated in VOD pathology. Key cellular components include the plasma membrane, nucleus, cytoplasm, and extracellular matrix. This analysis helps localize critical molecular interactions within VOD-affected cells.
The MF category described the specific biochemical activities of gene products, such as enzymatic functions, binding affinity, and signal transduction. Examining MFs enabled us to elucidate the primary functional roles of shared genes at a molecular level.
Network construction and Hub gene identification
Networks of common genes were constructed using Cytoscape software. The CytoHubba plugin [15] was employed to identify hub genes within these networks. A network comprising 87 nodes and 410 edges was constructed for the 159 genes associated with the overexpressed miRNAs, enabling the identification of potential key players in the pathogenesis of VOD.
Findings
Identification of differentially expressed miRNAs in VOD samples and target gene prediction
Using the GSE164635 dataset, which includes 30 samples (nine control samples, nine VOD patient samples, and 12 samples with other liver diseases), we identified four differentially expressed miRNAs (DEmiRNAs) through stringent filtration criteria (logFC > +1 or < -1 and adj.P.Val < 0.05). The identified DEmiRNAs were hsa-miR-194-5p, hsa-miR-148a-3p, hsa-miR-342-3p, and hsa-miR-150-5p. To validate our findings, we confirmed the association of these miRNAs with relevant diseases using the Human MicroRNA Disease Database (HMDD) [16], supporting the potential clinical relevance of these miRNAs in VOD. Notably, hsa-miR-194-5p was highlighted among the identified miRNAs, further underscoring its significance (Figure 1A).
Target gene prediction for these miRNAs was performed using the multiMiR package in R, with data sourced from the mirTarBase and Tarbase databases. For the two upregulated miRNAs (hsa-miR-194-5p and hsa-miR-148a-3p), 159 common target genes were identified. Similarly, 74 common target genes were identified for the two downregulated miRNAs (hsa-miR-342-3p and hsa-miR-150-5p). The overlap of the target genes for these miRNAs was depicted in Venn diagrams (Figure 1B and 1C).

Figure 1. Venn diagram illustrating the shared miRNAs and predicted target genes.
A. Intersection of the four identified differentially expressed miRNAs (DEmiRNAs) and their confirmed associations with relevant diseases from the Human miRNA Disease Database (HMDD). B. Overlap of target genes predicted for the upregulated miRNAs (hsa-miR-194-5p and hsa-miR-148a-3p), showing 159 common targets. C. Overlap of target genes for the downregulated miRNAs (hsa-miR-342-3p and hsa-miR-150-5p), indicating 74 common targets
Gene ontology and pathway analysis of identified target genes
For the 159 target genes of the upregulated miRNAs (hsa-miR-194-5p and hsa-miR-148a-3p), pathway enrichment and GO analyses were conducted using Funrich 3.1.3 software. The identified target genes were significantly enriched in several biological pathways, including the VEG and VEGFR signaling network, plasma membrane estrogen receptor signaling, glypican 1 network, proteoglycan syndecan-mediated signaling events, glypican pathway, and integrin family cell surface interactions (Figure 2A). In terms of BPs, the enriched categories included gene silencing, morphogenesis, RNA metabolism, epigenetic regulation of gene expression, cell communication, and signal transduction (Figure 2B). Cellular component analysis revealed significant enrichment in the RNA-induced silencing complex, cytosol, micro-ribonucleoprotein complex, lysosomes, exosomes, and the cytoplasm (Figure 2C). The enriched terms for MFs included GTPase activity, receptor signaling protein serine/threonine kinase activity, MHC class II receptor activity, MHC class I receptor activity, protein tyrosine phosphatase activity, and DNA methyltransferase activity (Figure 2D).

Figure 2. Gene ontology and pathway analysis of identified target genes for upregulated miRNAs. The most significant terms for biological process (BP), cellular component (CC), molecular function (MF), and biological pathway (Bipath) were identified and visualized using R software. Terms with a p-value and adjusted p-value<0.05 were considered significant
Gene-miRNA interaction for downregulated miRNAs
Similarly, for the 74 target genes of the downregulated miRNAs (hsa-miR-342-3p and hsa-miR-150-5p), pathway enrichment and GO analyses provided significant insights into the molecular mechanisms affected by these miRNAs. The biological pathways enriched for these target genes included validated transcriptional targets of AP1 family members Fra1 and Fra2, hypoxic and oxygen homeostasis regulation of HIF-1-alpha, the E2F transcription factor network, the HIF-2-alpha transcription factor network, the HIF-1-alpha transcription factor network, and the regulation of retinoblastoma proteins (Figure 3A). The BPs enriched in these target genes included transcription, lipid metabolism, protein folding, regulation of enzyme activity, vesicle docking, and regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolism (Figure 3B). Cellular component analysis revealed significant enrichment in the nodes of Ranvier, nuclear centromeric heterochromatin, protein complexes, membranes, endoplasmic reticulum membranes, and nuclei (Figure 3C). For molecular function, the enriched terms included receptor signaling protein tyrosine phosphatase activity, phosphatase regulator activity, kinase binding, DNA methyltransferase activity, protein tyrosine phosphatase activity, and transcription factor activity (Figure 3D).

Figure 3. Gene ontology and pathway analysis of identified target genes for down-regulated miRNAs
Gene-miRNA interaction networks
Gene-miRNA interaction for up-regulated miRNAs: Gene-miRNA interaction networks for upregulated and downregulated miRNAs were constructed using Cytoscape. For the upregulated miRNAs, the network revealed interactions with key genes, such as AGO2, CDKN1A, HSP90AA1, HSPA4, EP300, IGF1R, MYC, SMAD2, DICER1, and IL10. This network (Figure 4A) highlighted the central roles of these genes in various cellular processes and pathways.

Figure 4. Visualization of miRNA-target gene interaction
Gene-miRNA interaction for down-regulated miRNAs: For the downregulated miRNAs, the interaction network identified significant genes, including EEF2, IGF1R, EP300, CCN2, DNMT1, SREBF1, CANX, ZEB1, SP1, and JUN. This network (Figure 4B) underscores the involvement of these genes in transcriptional regulation, lipid metabolism, protein folding, and other functions (Figure 4B).
These comprehensive analyses provide valuable insights into the molecular mechanisms underlying VOD and suggest potential biomarkers and therapeutic targets for improving the diagnosis and treatment of this condition.
Discussion
This study aimed to identify novel blood biomarkers associated with veno-occlusive disease through bioinformatics analysis to improve early diagnosis and treatment outcomes. VOD is a severe condition characterized by the blockage of small hepatic veins. It primarily occurs after chemotherapy and radiation, particularly following HSCT. VOD is associated with various causes, including obesity and underlying diseases such as sickle cell disease, certain leukemias, and other hematological disorders. The functions of microRNAs (miRNAs) are highly relevant in numerous physiological and pathological processes, including inflammation, liver fibrosis, and cellular stress responses, all of which are crucial in the development and progression of VOD. In this session, we will discuss the miRNA/gene pathways involved in the mechanisms of action related to VOD.
Our results demonstrated interactions between the upregulated miRNAs (hsa-miR-194-5p and hsa-miR-148a-3p) and key genes, including AGO2, CDKN1A, HSP90AA1, HSPA4, EP300, IGF1R, MYC, SMAD2, DICER1, and IL10 in VOD. A recent study found that patients and rats with pyrrolizidine alkaloid-induced hepatic sinusoidal obstruction syndrome (HSOS) exhibit overexpression of three miRNAs: miR-148a-3p, miR-362-5p, and miR-194-5p [12]. AGO2, a key component of the RNA-induced silencing complex (RISC), along with DICER1, which is an essential enzyme for the production of microRNAs (miRNAs), plays a critical role in regulating gene expression [17].
The insulin-like growth factor 1 receptor (IGF1R) is a transmembrane receptor that is crucial for growth and development. It mediates the effects of IGF-1, which is involved in various cellular processes, including proliferation, differentiation, and survival. IGF1R signaling is particularly important for liver function and has implications for various diseases, including cancer and metabolic disorders [18]. HSP90AA1, which encodes a molecular chaperone, and HSPA4, a heat shock protein, are essential in various cancers and diseases. HSP90AA1 promotes inflammation in the liver, potentially contributing to liver fibrosis [19, 20]. SMAD2 serves as a key mediator of the transforming growth factor beta (TGF-β) signaling pathway, which is critical in liver fibrosis. TGF-β is known to promote the activation of hepatic stellate cells (HSCs), leading to collagen deposition and fibrosis, which can exacerbate VOD [21, 22].
IL-10 is a potent anti-inflammatory cytokine that plays a protective role in various liver conditions, including VOD. It can inhibit the production of pro-inflammatory cytokines, thereby reducing inflammatory responses and tissue damage [23]. CDKN1A, also known as P21, is associated with cellular senescence and the cell cycle and is implicated in liver fibrosis and inflammation. Elevated CDKN1A expression has been observed in multiple cancer types after chemotherapy and radiotherapy [24].
EP300 (E1A-binding protein p300) is a transcriptional coactivator that plays a crucial role in regulating gene expression. It is involved in several cellular processes, including cell proliferation, differentiation, and apoptosis. EP300 is known for its histone acetyltransferase activity, which modifies chromatin and facilitates gene transcription by recruiting various transcription factors and RNA polymerase II to target genes [25]. Research suggests that EP300 inhibition can attenuate fibrosis-related molecular processes, including extracellular matrix deposition, inflammation, and epithelial-to-mesenchymal transition [26].
MYC is a family of regulatory genes that encode transcription factors involved in various cellular processes, such as cell proliferation, growth, differentiation, and apoptosis. MYC plays a significant role in oncogenesis and is dysregulated in various cancers. Aberrant MYC expression can lead to increased cell proliferation and resistance to apoptosis, contributing to tumor progression and metastasis [27].
Eukaryotic elongation factor 2 (EEF2) is a critical protein involved in protein synthesis (translation) within eukaryotic cells. It plays a vital role in the elongation phase of translation, facilitating the movement of the ribosome along the messenger RNA (mRNA) strand as amino acids are added to the growing polypeptide chain [28].
To date, no studies have suggested a connection between VOD and the expression of MYC, EEF2, or EP300. Nevertheless, these factors may influence pathways associated with VOD.
Furthermore, we established a network linking the downregulated hsa-miR-342-3p and hsa-miR-150-5p to their targets EEF2, IGF1R, EP300, CCN2, DNMT1, SREBF1, CANX, ZEB1, SP1, and JUN. In hepatocellular carcinoma (HCC), miR-342-3p acts as a potent tumor suppressor [29]. Additionally, miR-342-3p influences HCC cell proliferation and has potential as a target for investigating therapeutics [30].
Hsa-miR-150-5p is a microRNA (miRNA) that has been studied in relation to lipid metabolism. Various studies suggest that it may play a role in hepatic lipid homeostasis by influencing lipid accumulation in the liver [31]. Another study showed that miR-150-5p exhibits opposite regulation and function in HSCs and hepatocytes during liver fibrosis [32].
Cellular communication network 2 (CCN2) is involved in cell migration, angiogenesis, and tissue repair. Studies have reported that CCN2 is associated with various aspects of lipid metabolism, including adipogenesis, lipolysis, lipid synthesis, and lipid oxidative stress [33, 34].
DNMT1, or DNA methyltransferase 1, is involved in DNA methylation, a key epigenetic mechanism. Nutrient-rich diets may upregulate DNA methyltransferases, leading to hypermethylation of the Klb promoter and downregulation of Klb expression, which impairs fatty acid oxidation and contributes to the development of NAFLD in male C57BL/6J mice [35].
SREBF1, also known as sterol regulatory element-binding transcription factor 1, regulates genes involved in lipid metabolism. SREBPs can participate in the development of liver fibrosis through a lipid-independent pathway. However, SREBP-1c can inhibit the activation of HSCs and liver fibrosis by downregulating TGF-β, SMAD3, and AKT [36].
Calnexin (CANX) is a protein involved in protein folding in the endoplasmic reticulum. Recent studies suggest that calnexin does more than assist with protein folding; it also interacts with other proteins involved in signaling and lipid metabolism. One of these proteins, PTP-1B, is attached to the ER membrane, similar to DGAT2. The connection between PTP-1B and the ER relies on calnexin [37].
ZEB1, or zinc finger E-box binding homeobox 1, is a transcription factor involved in cell differentiation and epithelial-mesenchymal transition. SP1, or Specificity Protein 1, is a transcription factor implicated in various BPs, including cell growth and apoptosis. Research has shown that miR-124 targets specificity protein 1 (Sp1), an important gene for liver lipid metabolism [38, 39].
JUN is a transcription factor involved in cell growth, apoptosis, and differentiation. Gao et al. reported that in the presence of non-esterified fatty acids (NEFA), activation of the inositol-requiring enzyme 1α and c-Jun N-terminal kinase axis contributes to intracellular lipid accumulation in calf hepatocytes [40].
Continuous research is essential for further exploration of the roles of miRNAs in VOD and for uncovering their potential clinical applications. This study highlights the significant role of miRNAs in VOD, suggesting potential biomarkers for early diagnosis and intervention. Identifying these biomarkers could lead to improved screening methods for high-risk populations, allowing for timely therapeutic strategies that may mitigate the severity of VOD. Moreover, understanding the molecular mechanisms underlying VOD can inform preventive measures, particularly in patients undergoing HSCT or those with predisposing conditions. By targeting specific miRNA pathways, future therapies could not only treat existing conditions but also prevent the onset of VOD in susceptible individuals. Further investigation is needed to identify specific miRNAs. Animal models are being employed to examine the impact of specific miRNAs on the pathophysiology of the disease. Additionally, advancements in genomics may pave the way for clinical studies that establish a correlation between miRNA profiles and VOD outcomes in patients. This field could potentially lead to the discovery of innovative biomarkers for diagnosis and the development of novel therapeutic approaches for managing VOD.
While our findings contribute to the understanding of miRNA pathways in VOD, several limitations should be considered. The study’s reliance on specific models may not fully represent the complexity of VOD in human patients. Additionally, the sample size for some miRNA analyses was limited, which could affect the generalizability of the findings. Further research with larger cohorts and diverse populations is necessary to validate these results and explore the clinical implications of the identified biomarkers.
Conclusion
The pathophysiology of VOD is greatly influenced by microRNAs, which play a crucial role in regulating inflammation, fibrosis, endothelial function, and cellular survival.
Acknowledgments: We are sincerely grateful for the support of the professors and staff of Islamic Azad University, Damghan Branch during the conduct of this research. We also acknowledge the contributions of the researchers who provided access to the GSE164635 dataset and the developers of the R packages used in our analysis. Special thanks to our colleagues for their valuable views and feedback during the study
Ethical Permissions: This study was conducted in accordance with the ethical standards of Islamic Azad University, Damghan branch. Ethical approval was obtained from the university's Institutional Review Board (IRB). All patient data were anonymized to ensure confidentiality and conform to ethical guidelines for research involving human subjects.
Conflicts of Interests: The authors reported no conflicts of interests.
Authors' Contribution: Mohammadi P (First Author), Introduction Writer/Main Assistant Researcher/Discussion Writer (60%); Noruzinia M (Second Author), Methodologist/Original or Assistant Researcher/Discussion Writer/Statistical Analyst (20%); Ebadi M (Third Author), Assistant Researcher (10%); Ghoraeian P (Fourth Author), Discussion Writer/Statistical Analyst (10%)
Funding/Support: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Article Type: Original Research |
Subject:
Health Communication
Received: 2024/09/7 | Accepted: 2024/10/20 | Published: 2024/10/30
Received: 2024/09/7 | Accepted: 2024/10/20 | Published: 2024/10/30
References
1. Richardson PG, Ho VT, Cutler C, Glotzbecker B, Antin JH, Soiffer R. Hepatic veno-occlusive disease after hematopoietic stem cell transplantation: Novel insights to pathogenesis, current status of treatment, and future directions. Biol Blood Marrow Transplant. 2013;19(Suppl 1):S88-90. [Link] [DOI:10.1016/j.bbmt.2012.10.023]
2. Bonifazi F, Barbato F, Ravaioli F, Sessa M, Defrancesco I, Arpinati M, et al. Diagnosis and treatment of VOD/SOS after allogeneic hematopoietic stem cell transplantation. Front Immunol. 2020;11:489. [Link] [DOI:10.3389/fimmu.2020.00489]
3. Dalle JH, Giralt SA. Hepatic veno-occlusive disease after hematopoietic stem cell transplantation: Risk factors and stratification, prophylaxis, and treatment. Biol Blood Marrow Transplant. 2016;22(3):400-9. [Link] [DOI:10.1016/j.bbmt.2015.09.024]
4. Richardson PG, Ho VT, Giralt S, Arai S, Mineishi S, Cutler C, et al. Safety and efficacy of defibrotide for the treatment of severe hepatic veno-occlusive disease. Ther Adv Hematol. 2012;3(4):253-65. [Link] [DOI:10.1177/2040620712441943]
5. Lewis TC, Cortes J, Altshuler D, Papadopoulos J. Venous thromboembolism prophylaxis: A narrative review with a focus on the high-risk critically ill patient. J Intensive Care Med. 2019;34(11-12):877-88. [Link] [DOI:10.1177/0885066618796486]
6. Cutler C, Kim HT, Ayanian S, Bradwin G, Revta C, Aldridge J, et al. Prediction of veno-occlusive disease using biomarkers of endothelial injury. Biol Blood Marrow Transplant. 2010;16(8):1180-5. [Link] [DOI:10.1016/j.bbmt.2010.02.016]
7. Fitzpatrick E, Dhawan A. Noninvasive biomarkers in non-alcoholic fatty liver disease: Current status and a glimpse of the future. World J Gastroenterol. 2014;20(31):10851-63. [Link] [DOI:10.3748/wjg.v20.i31.10851]
8. Ladero JM. Noninvasive evaluation of liver fibrosis in patients with chronic hepatitis C. Hepat Mon. 2011;11(9):698-700. [Link] [DOI:10.5812/kowsar.173543X.738]
9. Weng JT, Wu LC, Chang WC, Chang TH, Akutsu T, Lee TY. Novel bioinformatics approaches for analysis of high-throughput biological data. Biomed Res Int. 2014;2014:814092. [Link] [DOI:10.1155/2014/814092]
10. Oh JH, Craft JM, Townsend R, Deasy JO, Bradley JD, El Naqa I. A bioinformatics approach for biomarker identification in radiation-induced lung inflammation from limited proteomics data. J Proteome Res. 2011;10(3):1406-15. [Link] [DOI:10.1021/pr101226q]
11. Pritzker KP, Pritzker LB. Bioinformatics advances for clinical biomarker development. Expert Opin Med Diagn. 2012;6(1):39-48. [Link] [DOI:10.1517/17530059.2012.634797]
12. Wang X, Zhang W, Yang Y, Chen Y, Zhuge Y, Xiong A, et al. Blood microRNA signatures serve as potential diagnostic biomarkers for hepatic sinusoidal obstruction syndrome caused by Gynura japonica containing pyrrolizidine alkaloids. Front Pharmacol. 2021;12:627126. [Link] [DOI:10.3389/fphar.2021.627126]
13. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [Link] [DOI:10.1093/nar/gkv007]
14. Ru Y, Kechris KJ, Tabakoff B, Hoffman P, Radcliffe RA, Bowler R, et al. The multiMiR R package and database: Integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 2014;42(17):e133. [Link] [DOI:10.1093/nar/gku631]
15. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. CytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4(Suppl 4):S11. [Link] [DOI:10.1186/1752-0509-8-S4-S11]
16. Cui C, Zhong B, Fan R, Cui Q. HMDD v4.0: A database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2024;52(D1):D1327-32. [Link] [DOI:10.1093/nar/gkad717]
17. Pokornowska M, Milewski MC, Ciechanowska K, Szczepańska A, Wojnicka M, Radogostowicz Z, et al. The RNA-RNA base pairing potential of human Dicer and Ago2 proteins. Cell Mol Life Sci. 2020;77(16):3231-44. [Link] [DOI:10.1007/s00018-019-03344-6]
18. Gui R, Li W, Li Z, Wang H, Wu Y, Jiao W, et al. Effects and potential mechanisms of IGF1/IGF1R in the liver fibrosis: A review. Int J Biol Macromol. 2023;251:126263. [Link] [DOI:10.1016/j.ijbiomac.2023.126263]
19. Matthews I, Duong M, Parsons VL, Nozad B, Qizilbash N, Patel Y, et al. Burden of disease from shingles and post-herpetic neuralgia in the over 80 year olds in the UK. PLoS One. 2020;15(2):e0229224. [Link] [DOI:10.1371/journal.pone.0229224]
20. Choudhury A, Ratna A, Lim A, Sebastian RM, Moore CL, Filliol AA, et al. Loss of heat shock factor 1 promotes hepatic stellate cell activation and drives liver fibrosis. Hepatol Commun. 2022;6(10):2781-97. [Link] [DOI:10.1002/hep4.2058]
21. Xu F, Liu C, Zhou D, Zhang L. TGF-β/SMAD pathway and its regulation in hepatic fibrosis. J Histochem Cytochem. 2016;64(3):157-67. [Link] [DOI:10.1369/0022155415627681]
22. Dewidar B, Meyer C, Dooley S, Meindl-Beinker AN. TGF-β in hepatic stellate cell activation and liver fibrogenesis-updated 2019. Cells. 2019;8(11);1419. [Link] [DOI:10.3390/cells8111419]
23. Steen EH, Wang X, Balaji S, Butte MJ, Bollyky PL, Keswani SG. The role of the anti-inflammatory cytokine interleukin-10 in tissue fibrosis. Adv Wound Care. 2020;9(4):184-98. [Link] [DOI:10.1089/wound.2019.1032]
24. Manousakis E, Miralles CM, Esquerda MG, Wright RHG. CDKN1A/p21 in breast cancer: Part of the problem, or part of the solution?. Int J Mol Sci. 2023;24(24):17488. [Link] [DOI:10.3390/ijms242417488]
25. Kikuchi M, Morita S, Wakamori M, Sato S, Uchikubo-Kamo T, Suzuki T, et al. Epigenetic mechanisms to propagate histone acetylation by p300/CBP. Nat Commun. 2023;14:4103. [Link] [DOI:10.1038/s41467-023-39735-4]
26. Rubio K, Molina-Herrera A, Pérez-González A, Hernández-Galdámez HV, Piña-Vázquez C, Araujo-Ramos T, et al. EP300 as a molecular integrator of fibrotic transcriptional programs. Int J Mol Sci. 2023;24(15):12302. [Link] [DOI:10.3390/ijms241512302]
27. Scafuro M, Capasso L, Carafa V, Altucci L, Nebbioso A. Gene transactivation and transrepression in MYC-driven cancers. Int J Mol Sci. 2021;22(7):3458. [Link] [DOI:10.3390/ijms22073458]
28. Susorov D, Zakharov N, Shuvalova E, Ivanov A, Egorova T, Shuvalov A, et al. Eukaryotic translation elongation factor 2 (eEF2) catalyzes reverse translocation of the eukaryotic ribosome. J Biol Chem. 2018;293(14):5220-9. [Link] [DOI:10.1074/jbc.RA117.000761]
29. Komoll RM, Hu Q, Olarewaju O, Von Döhlen L, Yuan Q, Xie Y, et al. MicroRNA-342-3p is a potent tumour suppressor in hepatocellular carcinoma. J Hepatol. 2021;74(1):122-34. [Link] [DOI:10.1016/j.jhep.2020.07.039]
30. Zhao L, Zhang Y. miR-342-3p affects hepatocellular carcinoma cell proliferation via regulating NF-κB pathway. Biochem Biophys Res Commun. 2015;457(3):370-7. [Link] [DOI:10.1016/j.bbrc.2014.12.119]
31. Moraes VN, Queiroz AL, Martone D, Rodrigues JAL, Gomes MM, Salgado JÚnior W, et al. Relationship between the hsa miR 150-5p and FTO gene expression in white subcutaneous adipose tissue with overweight/obesity, lipid profile and glycemia. An Acad Bras Cienc. 2020;92(4):e20200249. [Link] [DOI:10.1590/0001-3765202020200249]
32. Chen W, Yan X, Yang A, Xu A, Huang T, You H. miRNA-150-5p promotes hepatic stellate cell proliferation and sensitizes hepatocyte apoptosis during liver fibrosis. Epigenomics. 2020;12(1):53-67. [Link] [DOI:10.2217/epi-2019-0104]
33. Yang A, Mottillo EP. Adipocyte lipolysis: From molecular mechanisms of regulation to disease and therapeutics. Biochem J. 2020;477(5):985-1008. [Link] [DOI:10.1042/BCJ20190468]
34. Liu B, Dai Z. Fatty acid metabolism in endothelial cell. Genes. 2022;13(12):2301. [Link] [DOI:10.3390/genes13122301]
35. Wang S, Zha L, Cui X, Yeh YT, Liu R, Jing J, et al. Epigenetic regulation of hepatic lipid metabolism by DNA methylation. Adv Sci. 2023;10(20):2206068. [Link] [DOI:10.1002/advs.202206068]
36. Li N, Li X, Ding Y, Liu X, Diggle K, Kisseleva T, et al. SREBP regulation of lipid metabolism in liver disease, and therapeutic strategies. Biomedicines. 2023;11(12):3280. [Link] [DOI:10.3390/biomedicines11123280]
37. Brandt C, McFie PJ, Vu H, Chumala P, Katselis GS, Stone SJ. Identification of calnexin as a diacylglycerol acyltransferase-2 interacting protein. PLoS One. 2019;14(1):e0210396. [Link] [DOI:10.1371/journal.pone.0210396]
38. Cheng Y, Huang L, Ping J, Chen T, Chen J. MicroRNA-199a-3p attenuates hepatic lipogenesis by targeting Sp1. Am J Transl Res. 2017;9(4):1905-13. [Link] [DOI:10.1016/S0168-8278(18)30909-7]
39. Lee S, Mardinoglu A, Zhang C, Lee D, Nielsen J. Dysregulated signaling hubs of liver lipid metabolism reveal hepatocellular carcinoma pathogenesis. Nucleic Acids Res. 2016;44(12):5529-39. [Link] [DOI:10.1093/nar/gkw462]
40. Gao W, Wang Y, Liu S, Li G, Shao Q, Zhang C, et al. Inositol-requiring enzyme 1α and c-Jun N-terminal kinase axis activation contributes to intracellular lipid accumulation in calf hepatocytes. J Dairy Sci. 2024;107(5):3127-39. [Link] [DOI:10.3168/jds.2022-23189]
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |






